MRC WIMM Prize PhD Studentships 2018
Posted by ecloke, on 30 October 2017
Closing Date: 15 March 2021

The MRC Weatherall Institute of Molecular Medicine (WIMM) has fully funded 4-year Prize PhD (DPhil) Studentships available to start in October 2018. These Studentships are open to outstanding students of any nationality who wish to train in experimental and/or computational biology.
The Institute is a world leading molecular and cell biology centre that focuses on research with application to human disease. It includes the recently opened MRC WIMM Centre for Computational Biology and houses over 500 research and support staff in 50 research groups working on a range of fields in Haematology, Gene Regulation & Epigenetics, Stem Cell Biology, Computational Biology, Cancer Biology, Human Genetics, Infection & Immunity. The Institute is committed to training the next generation of scientists in these fields through its Prize PhD Studentship Programme.
The fully funded studentships include a stipend of £18,000 per annum and cover University and College fees.
Further information on the studentships, how to apply, and the projects available can be found at:
http://www.imm.ox.ac.uk/wimm-prize-studentships-2018
Closing date for submission of applications: Monday, 8 January 2018, 12 noon UK time.
Interviews will take place the week commencing 22 January 2018.
Pure Computational Biology Project Leaders
Hashem Koohy – Machine-learning in gene function, transcription regulation and immunology
Ed Morrissey – Quantitative biology of cell fate
Aleksandr Sahakyan – Regulatory chromosomal domains and genome architecture
Supat Thongjuea – Computational biology of single-cell transcription and gene regulation
Molecular and Cell Biology Project Leaders
Ahmed Ahmed – Experimental therapeutics
Chris Babbs – Causes of congenital anaemia
Oliver Bannard – B cell biology
Andrew Blackford – DNA damage and disease
Walter Bodmer – Colorectal cancer, stem cells, differentiation & drug response
Marella De Bruijn – Developmental haematopoiesis
Zam Cader – Stem cell neurological disease models
Vincenzo Cerundolo – Tumour immunology, vaccine strategies
David Clynes – DNA damage, repair and cancer
Simon Davis – T-cell biology
Hal Drakesmith – Iron and infection
Christian Eggeling – Super-resolution microscopy in immunology
Ben Fairfax – Inflammation, genetics and cancer therapeutics
Marco Fritzsche – Biophysical immunology
Lars Fugger – Multiple sclerosis
Tudor Fulga – MicroRNAs in development and disease
Richard Gibbons – Chromatin, epigenetics & transcription
Anne Goriely – De novo mutations and human disease
Doug Higgs – Gene regulation and epigenetics
Ling-Pei Ho – Lung immunology
Georg Hollander – T cell development and thymus organogenesis
David Jackson – Lymphatic trafficking in inflammation and cancer
Peter McHugh – DNA repair
Adam Mead – Normal and leukaemic haematopoietic stem cell biology
Claus Nerlov – Tissue stem cell genetics
Graham Ogg – Translational skin research
Catherine Porcher – Transcription factors and blood development
Jan Rehwinkel – Innate detection of viruses
Irene Roberts – Trisomy 21, haematopoiesis and leukaemia
Tatjana Sauka-Spengler – Neural crest gene regulatory networks
Alison Simmons – Innate immunity & Crohn’s disease
Alain Townsend – Influenza and ebola, vaccination and treatment
Paresh Vyas – Leukaemic stem cells
Andrew Wilkie – Sperm and craniofacial mutations
![]()


 (No Ratings Yet)
(No Ratings Yet)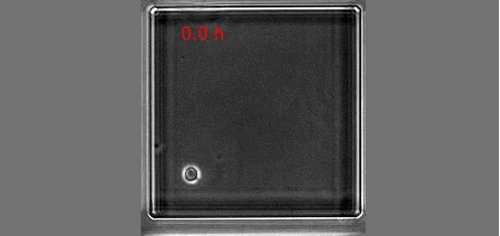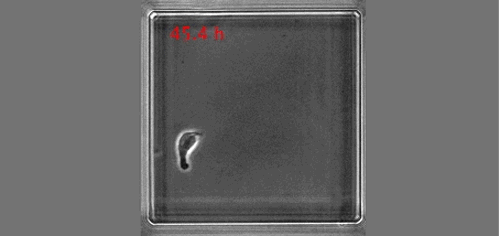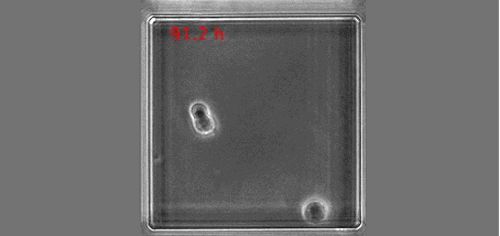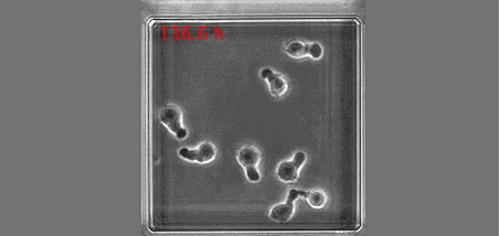
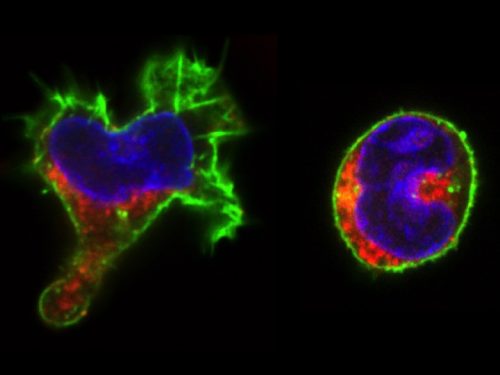
 (1 votes)
(1 votes)
 Christiana Ruhrberg studied Biology at the Justus Liebig University (Giessen, Germany), and obtained her first class Diploma/MSc degree in 1992. After taking on an MSc/research assistant position at the University of Sussex for two years to elucidate genetic changes in ovarian cancer, she moved to Imperial College London to work for another two years to study the genomic organisation of the gene-rich human ‘surfeit’ locus. She then carried out her PhD project in the laboratory of Fiona Watt at the Imperial Cancer Research Fund (1994-97) where she identified and described the function of the envoplakin and periplakin genes. Her postdoctoral research in the laboratories of Robb Krumlauf at the National Institute for Medical Research in London (1997-1999) was funded by a MRC postdoctoral training fellowship and dedicated to studying the role of Hoxa1, Hoxb1 and Hoxb2 during hindbrain development. During her second postdoc with David Shima at the Imperial Cancer Research Fund in London (2000-2002), she was funded by a ICRF fellowship and worked on VEGF-A-mediated blood vessel branching. Having received an MRC Career Development Award in 2003, she became an independent investigator at University College London’s Institute of Ophthalmology studying links between vascular and neuronal development, with particular focus on the roles of VEGF and SEMA3A signalling during facial nerve and blood vessel formation. Staying at that same institute, she was appointed Lecturer in 2007, promoted to Reader in 2008 and then full Professor in 2011. Christiana has an excellent record in obtaining research funding, as illustrated by a Wellcome Trust Junior Investigator Award in 2011 and a Wellcome Trust Investigator Award in 2016 for her research on vascular biology and neurovascular interactions.
Christiana Ruhrberg studied Biology at the Justus Liebig University (Giessen, Germany), and obtained her first class Diploma/MSc degree in 1992. After taking on an MSc/research assistant position at the University of Sussex for two years to elucidate genetic changes in ovarian cancer, she moved to Imperial College London to work for another two years to study the genomic organisation of the gene-rich human ‘surfeit’ locus. She then carried out her PhD project in the laboratory of Fiona Watt at the Imperial Cancer Research Fund (1994-97) where she identified and described the function of the envoplakin and periplakin genes. Her postdoctoral research in the laboratories of Robb Krumlauf at the National Institute for Medical Research in London (1997-1999) was funded by a MRC postdoctoral training fellowship and dedicated to studying the role of Hoxa1, Hoxb1 and Hoxb2 during hindbrain development. During her second postdoc with David Shima at the Imperial Cancer Research Fund in London (2000-2002), she was funded by a ICRF fellowship and worked on VEGF-A-mediated blood vessel branching. Having received an MRC Career Development Award in 2003, she became an independent investigator at University College London’s Institute of Ophthalmology studying links between vascular and neuronal development, with particular focus on the roles of VEGF and SEMA3A signalling during facial nerve and blood vessel formation. Staying at that same institute, she was appointed Lecturer in 2007, promoted to Reader in 2008 and then full Professor in 2011. Christiana has an excellent record in obtaining research funding, as illustrated by a Wellcome Trust Junior Investigator Award in 2011 and a Wellcome Trust Investigator Award in 2016 for her research on vascular biology and neurovascular interactions.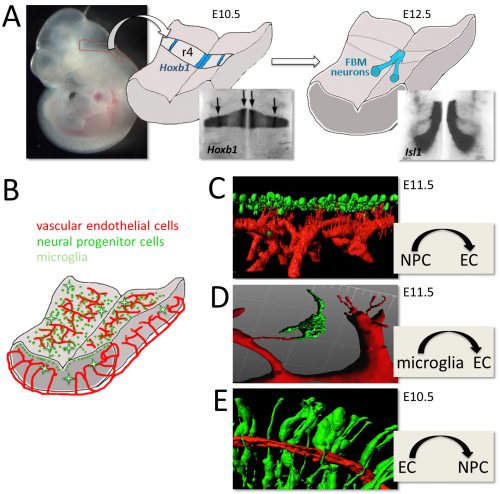
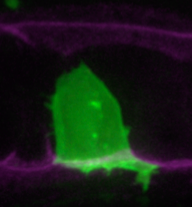
 Once upon a time a genetic screen identified a signalling pathway that caused Drosophila melanogaster embryos to develop a ‘lawn’ of denticles rather than forming them only at parasegment boundaries. Thus the so-called Hedgehog signaling pathway was born (
Once upon a time a genetic screen identified a signalling pathway that caused Drosophila melanogaster embryos to develop a ‘lawn’ of denticles rather than forming them only at parasegment boundaries. Thus the so-called Hedgehog signaling pathway was born (