Glia lead the way for pioneers to build the brain neuropil in C. elegans
Posted by Georgia Rapti, on 20 October 2017
It’s all about the wires. But what about the glue?
Networks make us who we are. I am not talking about social networks but about neural networks that define how we perceive the world and how we act. For a century, neuroscientists have sought to understand functions of neural networks in condition and how such networks are established in the first place still remains an active area of research. Neuronal networks consist of a large set of wires that need to be connected and organized in defined, functional ways, similarly to telecommunication wiring pervading our cities or like the motherboards of our modern computers. To work, signal carriers of such networks must be of appropriate bandwidth, with proper paths, and connected in a specific way. Likewise, in neuronal networks, cell fate must be specified, neurons must migrate to proper locations, grow and navigate their axon processes through specific paths and generate well-defined synaptic connections.
In the 1990’s, the “Decade of the Brain,” as declared by the United States Congress, developmental neuroscientists made extraordinary progress in understanding cell fate determination, axon guidance and synaptogenesis. Seminal studies identified the principal families of guidance molecules and established major paradigms, such as the role of floor plate Netrin, a secreted guidance cue, in commissural axon guidance 1,2. At the same time while some principles were established others were revisited. The non-neuronal cells of the nervous system, called glia (from the Greek γλία/ γλοία for glue), which as their name implies were long though to provide a passive substrate for neuronal growth, were realized to play active roles in neuronal physiology and function. Glia were first acknowledged to be neuron’s best friend, housekeeper, insulator, nurse 3,4and the list of glial roles expanded to include roles of glia as neural progenitors, tracts for migratory neurons and synaptic plasticity facilitators. Despite this progress, today’s classical neuroscience textbooks count only a handful of mentions of glia and their extensive roles. When I was an undergraduate, one would be lucky to attend a seminar on glial biology by one of the few scientists of the field.
I discovered the extensive glia literature when planning my post-doctoral research path. Focusing on the unknowns of neural development, I realized the enormous potential of the view that glia can actively regulate neuronal development and physiology. I was a descendant of the C. elegans research community, which I joined because of my first excitement about the C. elegans short life cycle and its recovery after freezing (allowing easy storage of strains for decades), but mainly for the ease of genetics, molecular biology and functional studies, possible at single-cell resolution. Yet, glial studies in this animal were few and far between. C. elegans axons are not myelinated by glia and glia were not shown to give rise to neurons nor to appose the neuromuscular junctions I studied during my PhD. In fact the presence of glia in C. elegans was not even clearly accepted. Then, midway during my PhD, Shai Shaham, my post-doc supervisor to-be, demonstrated that the so-called “support cells” of my favorite nematode are, in fact, glia 5, and soon after his lab demonstrated glial roles in neuronal ensheathment, dendrite shape and axon extension 6–8. Exciting! So many glial functions to be uncovered in C. elegans glia!
Yet, why would one study glial functions in C. elegans, where glial cells are limited to the 1/6 of the total neuron number, axons are non-myelinated and and microglia-like immune-specialized cells do not exist? For one, C. elegans has proven so useful for understanding the basic biology common to so many living things, that there was no reason to believe glia would be an exception! From a technical standpoint, most C. elegans neurons are born and grow independently of glia division and trophic support, thus perturbing glia does not cause neuronal death. In other systems, glial roles in neuron survival and support hamper attempts to address the active roles of glia in vivo in the nervous system. Finally, unlike most other settings, C. elegans glia can be studied in vivo, at single-cell resolution, using facile genetics, an invariant cell lineage and embryonic morphogenesis progresses in a transparent egg laid free in the environment. Moreover, live imaging of the same exact cell over and over again is possible in C. elegans. Who wouldn’t want to study glia in this setting!
The most complex circuit of the most well studied worm, and the forgotten cells
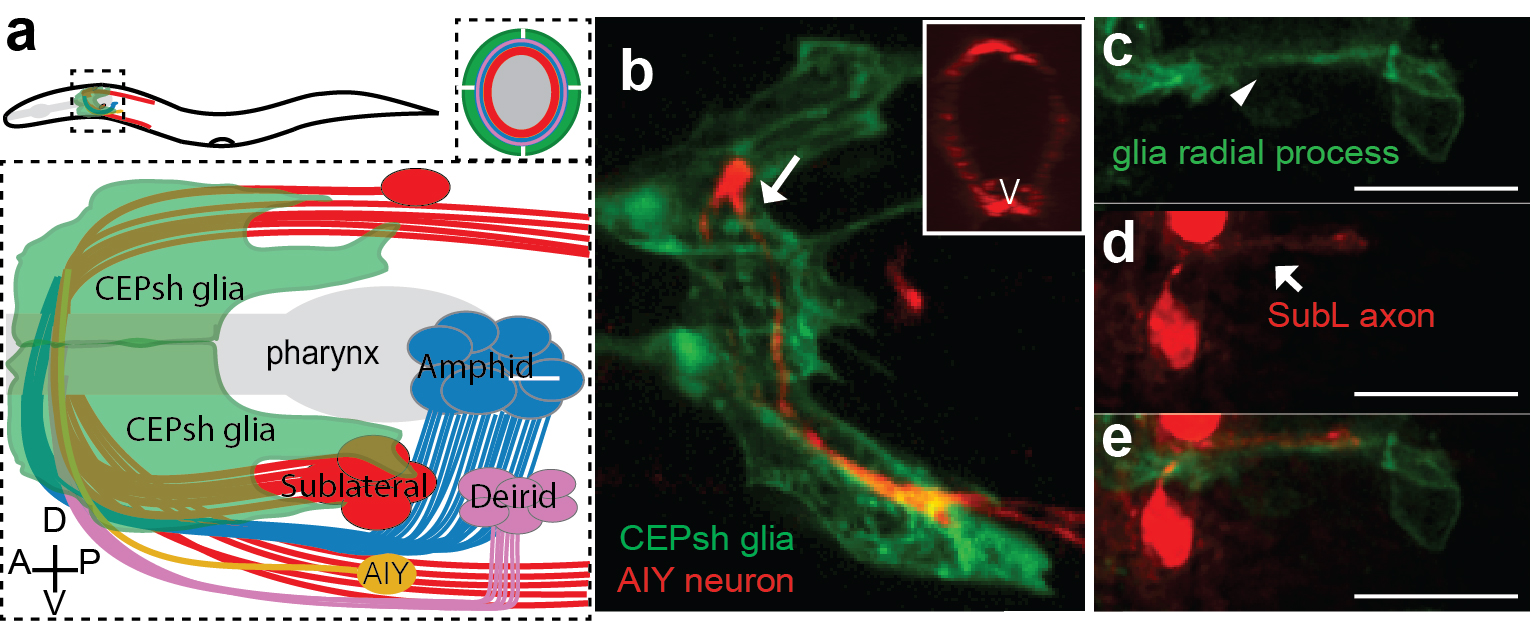
Armed with these thoughts, I joined Shai’s lab, decided to study developmental functions of glia, focusing on glial roles in the formation of the C. elegans brain-like neuropil, the nerve ring. The nerve ring (NR) consists of ~180 axons, presents the majority of synapses and interneuron connections in the animal, and is enveloped in its final form by four CEPsh glia, with astrocyte-like terminal morphology and molecular content (Figure 1; Katz & Shaham, personal communication, 9). CEPsh glia express Netrin in early embryogenesis 10. Moreover, ablation of their precursors results in more profound NR defects than those of Netrin null mutants, suggesting unknown glia roles in NR assembly 7,11. Thus, I was interested to focus on glial roles in NR assembly, using biased approaches.
A few weeks into the project, writing for fellowship applications, I was struck by an important realization. While C. elegans was an important model for axon guidance cue discovery 10,12,13 and for studies of axon growth and fasciculation 12–16, how its most complex neuropil is formed during embryogenesis was subject to extensive speculation but was not yet described in detail. Only a handful of studies focused on NR structure, axon guidance or positioning 7,11,13,17, and many important questions remained unanswered. When and where does NR assembly start in the embryo? How and when do NR components enter the structure? What are the first axons to pioneer the NR and do they have functional roles in guiding later “follower axons”? Do the CEPsh glia extend early processes to define the structure or do they only enclose the structure in later embryogenesis, after axons have fasciculated, to protect them from the mechanical forces during embryonic elongation? Is NR integrity regulated only by Netrin-expressing glia (ventral CEPsh glia) or also by the non-expressing ones (dorsal CEPsh glia)? Which molecules, besides Netrin, may function from glia or from early neurons to address NR formation? Are there molecules that control specifically axon guidance, independently from neuronal migration and axon-growth initiation? I was surprised that so many questions remained open in C. elegans, a model system well appreciated for embryonic studies, tractable neuronal identities and single-cell resolution. Were the answers to these questions trivial, and not worth exploring? Or, as I have heard some neuroscientists say, is neural development mostly explained, and only the dynamic plasticity of neuronal circuits remains to be understood? Was I doomed to engage in a problem solved in other organisms, to provide the C. elegans version “merely” for the record?
I had to really understand the state of affairs in the field, and in reading the literature realized more and more how little was known. Not only in C. elegans, but at all. Indeed, my attempts to prepare introductory slides for my early scientific presentations did not prove easy. Transcriptional and morphogen signaling pathways for neuronal specification were defined in detail 18–20 and growth cone morphology distinguished pioneer and follower axons 21–23. However, how neuronal fates dictated circuit assembly initiation through a series of comprehensible axon-guidance events was, with few exceptions, not at all clear. The molecular identities of pioneer axons and the molecular mechanisms driving their functional roles remained understudied. Glia were known to form transient structures associated with axonal bundles and to express guidance cues 24. However, their functional roles in axon guidance independently of neuronal survival, growth, migration and their molecular interactions with pioneer axons appeared unresolved 25–27. I was reminded of a century-old Cajal’s quote: “What functional significance can be attributed to the neuroglia? Unfortunately, the present state of science does not allow to answer this important question but through more or less rational conjectures.” (Ramón y Cajal, 1899).
First things first: CEPsh glia and SubL pioneers initiate the nerve ring neuropil
I decided to start from scratch. Instead of biasing my studies to focus on glia, I decided to observe and image NR development, and let nature be my guide. The post-embryonic NR contains two major commissures, the amphid and deirid commissures composed primarily of sensory neuron axons 17. The posterior part of the amphid commissure is also referred to as the sublateral (SubL) commissure, since its axons extend out of the NR to populate the ventral sublateral cords. Since the amphid commissure is the largest characterized bundle populating the NR we hypothesized that this may be the one growing out first. I started imaging amphid-neuron axons in the beginning of morphogenesis, initially by using recently published reporters 28. Imaging bundled membranes using the by Pdyf-7:: membrane-GFP reporter, or single axons, by photoconversion of specific neurons in animals expressing Pdyf-7::kaede, revealed axon growth only after 400 min of embryogenesis (known as the comma stage). Yet, electron microscopy (EM) images acquired by Yun Lu in the lab, showed that the pharyngeal primordium was surrounded by a thick layer of axons, corresponding to the early NR, already at 440min of embryogenesis (1.5-fold embryonic stage). So many axons were present already at 440min, making us wonder whether amphid neurons were truly early NR components.
I found myself, again, going one step back, realizing that I needed a more comprehensive effort to visualize as many embryonic neurons as I could. Since C. elegans has been a valuable model system for studies of neuronal specification and diversity 29, I was not too worried about finding neuron-subtype specific markers. I started to evaluate known neuronal markers. This screen left me with a handful of useful markers, and a couple of important take-home lessons. First, markers with post-embryonic expression restricted to specific neurons are usually more broadly expressed during embryonic morphogenesis. The C. elegans research community would, therefore, benefit greatly from characterizing expression patterns also during embryonic morphogenesis. Indeed, several labs today focus on expression profiling of C. elegans embryos 30,31, a blessed endeavor. This realization, that sparse, cell-specific embryonic reporters were few and far between, also drove the development in Shai’s lab of a method for labeling individual C. elegans embryonic cells without cell-specific drivers 32. Second, the willingness of the C. elegans research community to share reagents, published dozens of years ago or others unpublished, to help me with my project, proved the supportive nature of this community, something that became rare in biological research.
I was now armed with reporters for many axons populating distinct NR commissures. By performing in vivo live embryonic timelapses of process growth into the NR, I saw that axons of the sublateral bundle grow early to define the NR neuropil while other commissures, amphid and deirid, were established in an orderly fashion later, with sequential axon growth occurring even within commissures (Figure 2a-b, 9). After months of sampling and imaging live embryos, I could not sustain my excitement when we also identified two bundles of similar position and composition in EM images of comma-stage embryos, taken by Yun Lu in the lab. I kept going back to analyze those EM images; yes, there were these two bundles, and no other multiprocess-bundles were obvious at that time, navigating to the NR presumptive position!
But what about the CEPsh glia? I had begun attempts to image CEPsh glia membrane growth the minute I joined Shai’s lab, but at the time no useful information could be gleaned, since no CEPsh-specific embryonic promoters were known. I decided to dissect regulatory elements of a couple of known CEPsh-expressing promoters, but this approach was not fruitful, as deletion analysis of cis-regulatory elements resulted in derepressed expression in non-glial cells. I finally decided to use a transgenic pan-glial marker, expressed from the onset of embryonic morphogenesis, and to follow stochastic segregation of the marker in my cells with mosaic analysis. Despite the method’s low throughput nature, I was able to visualize CEPsh membranes growing in the NR early and coalescing with growing SubL axons. Those early CEPsh membranes did not present the elaborate astrocyte-like endfeet seen post-embryonically but grow thin, non-branched processes reminiscent of vertebrate radial glia.
SubL pioneers and CEPsh populated the NR early! But who actually took the lead? Were SubL and CEPsh equally important in NR initiation or does one of the cell types guide the other? The answer came from ablation experiments I performed several months later. When specific CEPsh glia are ablated by expressing an apoptotic gene and following cell killing by mosaic analysis, pioneer axons grow ectopically, while follower axons can be short or mis-guided. Cell ablation of the SubL bundle perturbed follower axon pathfinding but CEPsh membrane morphology appeared normal. Thus, the most exciting scenario I could have imagined emerged: glia are not just bystander support cells maintaining the neuropil; they are necessary for initiating NR assembly. Moreover, our later studied revealed that CEPsh also cooperate with SubL pioneers to guide later NR components (Figure 2). This was an important realization. Although glia in other systems have been reported to appose axon bundles, functional analysis of glial roles in pioneer axon guidance was lacking, or was tested in settings where defects in neuronal viability or migration complicated the analysis. 26,27,33–35.
An “unsuccessful screen” & the first viable mutant with severe NR assembly defects
Defining a working model of cellular interactions for NR initiation was amazing, but not enough! Long devoted to unbiased genetics, like many C. elegans researchers, I decided to search for genes regulating NR formation through forward genetic screens. I screened tens of thousands of mutagenized worms under a compound microscope, recovering from the slides, back to bacteria-seeded plates, worms with abnormal axon trajectories. I started the screen well before my embryonic imaging uncovered the identities of early NR components. Back then I was thinking about neuropil formation, and selective fasciculation of axons. I hypothesized that fasciculation molecules, specific to neuron-subtypes, would ensure stereotypical interactions between axonal partners. I, therefore, designed screens for mutants in which two normally fasciculated axons, are properly guided but fail to adhere. My screens gave no viable mutants harboring the defects I was expecting. The reason for this remains unknown, but may suggest a different scenario of axon partnership based on the relative cell positioning of specific neurons at the time of their axon growth.
Regardless, in these screens I did find mutants with other NR axon defects. By now, I knew of the requirement of glia for NR assembly and decided to focus on mutants I found whose NR axon defects resembled those of glia-ablated animals. After cloning several previously-identified guidance genes with mild NR guidance defects, I decided to focus on a mutant with very severe NR defects, in which no mutations in the classical guidance genes were found by whole genome sequencing (WGS). And here started the painstaking tale of identifying the relevant gene. During my PhD, I mapped the first mutant of an uncharacterized gene without resorting to whole genome sequencing (WGS). It was difficult for me to understand why cloning my current mutant was so challenging, even with WGS data. The answer came when I proved in multiple ways that not one, but two mutations in two different unlinked genes were causal for the defects in the mutant. These mutations were synergistic, such that either single mutation caused only mild defects (5-15%) compared to the severe NR defect of the double mutant (>75%). To add to the complexity, one of the mutations was semi-dominant and subject to partial maternal rescue. After months of effort, I could finally announce that mutations in the Chimaerin gene chin-1, and the Furin gene kpc-1 were the cause of the mutant defects in NR axon guidance. When, in a joint group meeting, Shai, Leslie Vosshall, and Cori Bargmann commented on the low probability of recovering and identifying such a mutant, I felt that my hard work had really paid off. I was also filled with optimism that today’s ease of WGS, facilitating such complicated endeavors, will hopefully help identification of mutants that in previous decades would have been frozen and forgotten. This should allow researchers to choose their mutants of interest mainly based on interesting phenotypes and not how easy their identification is. For one, our mutant was definitely of interest; overall it showed ~80% navigation defects in any NR axon we imaged by optical means and a severe defective overall NR structure by electron microscopy of newly-hatched animals.
A wrong educated guess and the non-canonical functions of “old players”
The identities of the genes we found and previous studies in C. elegans or vertebrate nervous systems prompted me to favor cell-autonomous functions of these genes 36–38. I devoted several months to unsuccessfully prove such cell-autonomous roles of those in axon guidance. I was finally forced to consider the possibility of non-cell-autonomous roles. Rescue studies using combinations of cell-specific embryonic promoters finally revealed that the C. elegans Chimaerin and Furin act from glia to guide pioneer SubL axons and from both glia and pioneer SubL to guide follower axons. And they do so at the onset of NR assembly, by specifically affecting NR axon guidance, and not neuronal or glia survival, fate specification, migration or differentiation. To our knowledge, this is the first mutant affecting NR formation in such a specific way.
This mutant gave us an inroad in studying what turned out to be highly redundant signaling pathways for neuropil assembly initiation, a problem that has plagued genetic analysis of the process not only in C. elegans, but in vertebrates as well. Using the synergistic nature of Chimaerin and Furin mutants, we screened for mutations enhancing either single mutant and identified a network of conserved axon guidance cues redundantly regulating NR assembly. Importantly using our system of hierarchical NR assembly we could precisely define the cell-specific contributions of those factors. It turned out that knowing the site of action of these cues was crucial for modeling their actions. We demonstrated that C. elegans glia use distinct Netrin and Semaphorin signaling pathways to guide pioneer and follower axons respectively. We further showed glia and pioneer neurons together use the C. elegans Celsr/Flamingo homolog, FMI-1, to ensure proper navigation of follower axons into the neuropil. Moreover, our results support roles for Chimaerin and Furin in trafficking guidance cues, a novel combined function for this pair of proteins (Figure 2).
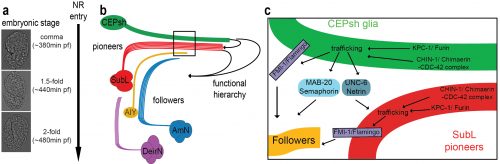
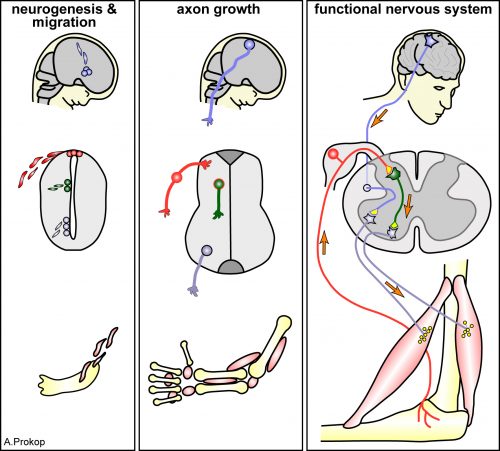
(a)The NR is assembled, orderly, from the comma till after the 2-fold embryonic stage. (b) CEPsh glia and pioneer SubL axons enter the path first, later followed by “follower” axons of commissures or non-commissural paths. This follows a functional hierarchy of CEPsh glia guiding SubL pioneers and cooperating with them to guide follower axons. (c) Molecular pathways function redundantly from CEPsh glia and SubL pioneers to drive NR assembly. Figure adapted from Rapti et al, 2017 (9).
In summary, our studies uncovered the initiating events of NR formation in C. elegans, identified key pioneer roles for glia, defined the identities of the first pioneer neurons that enter this brain-like neuropil, and uncovered a molecular framework governing axon guidance events early on. Our studies uncovered a genetic strategy for dissecting the highly redundant cellular and molecular interactions driving axon guidance, and defined a new role for two old players in guidance factor trafficking. This sounds like a lot of work. And it was. But really, it is just the beginning. There are so many questions left unanswered, and the logic of assembly is still not understood for most of the neurons entering the NR. How relevant these studies are to other systems also remains to be explored, but some exciting similarities have recently emerged (see below).
New ways of thinking about old molecules and established concepts
In retrospect, our findings did not provide a short, easy-to-digest, easy-to-sell story. Instead, by tackling a multifaceted problem without shying away from the complexity we took to heart Einstein’s quip that phenomena must be explained in the simplest possible terms, but no simpler. This is an important lesson- while there is great temptation to understand biology in well-packaged sound bites, life turns out usually to be a lot more complex. Oversimplifying can have the unwanted effect of preventing fundamental understanding of a process, and can lead to proclamations of entire scientific fields being “solved”, even when this is not the case. In our case, the benefit of revisiting old problems with unbiased approaches was the discovery of new concepts, like the new roles of previously known molecules and importantly the glia roles in initiating assembly.
Remarkably, as our manuscript was nearing publication, two papers were published that questioned the precise roles of Netrin in commissural axon guidance in vertebrates 39,40. While questioning the long-standing role of the floor plate in commissural axon guidance, they revealed important roles for Netrin derived from ventricular-zone neural progenitors that correspond to radial glia. These studies help put our discoveries in a much broader context. In our paper, we suggest that embryonic CEPsh glia resemble radial glia, both morphologically and molecularly, suggesting that vertebrate radial glia may guide pioneer axons to initiate circuit assembly, in addition to their recognized roles in neuronal migration. The recent Netrin papers greatly strengthen our hypothesis, and predict that the proteins and gene interactions we described in our paper are likely to exist in vertebrates as well.
This project taught me a valuable lesson: keep an open mind; you may be surprised.
And who doesn’t like surprises!?
This post is a comment on the paper: Rapti,G., Li, C., Shan,A., Lu,Y., Shaham,S. (2017). Glia initiate brain assembly through noncanonical Chimaerin–Furin axon guidance in C. elegans. nature neuroscience. 20, 1350-1360.
Georgia Rapti, PhD Shaham Laboratory The Rockefeller University, NYC
References
- Plachez, C. & Richards, L. J. Mechanisms of axon guidance in the developing nervous system. Curr. Top. Dev. Biol. 69, 267–346 (2005).
- Serafini, T. et al. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell 87, 1001–1014 (1996).
- Barres, B. a. New roles for glia. J. Neurosci. 11, 3685–3694 (1991).
- Vernadakis, A. Glia-neuron intercommunications and synaptic plasticity. Prog. Neurobiol. 49, 185–214 (1996).
- Shaham, S. Glia-neuron interactions in the nervous system of Caenorhabditis elegans. Curr. Opin. Neurobiol. 16, 522–528 (2006).
- Perens, E. A. & Shaham, S. C. elegans daf-6 encodes a patched-related protein required for lumen formation. Dev. Cell 8, 893–906 (2005).
- Yoshimura, S., Murray, J. I., Lu, Y., Waterston, R. H. & Shaham, S. mls-2 and vab-3 Control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Development 135, 2263–2275 (2008).
- Bacaj, T., Tevlin, M., Lu, Y. & Shaham, S. Glia are essential for sensory organ function in C. elegans. Science (80-. ). 322, 744–747 (2008).
- Rapti, G., Li, C., Shan, A., Lu, Y. & Shaham, S. Glia initiate brain assembly through noncanonical Chimaerin–Furin axon guidance in C. elegans. Nat. Neurosci. (2017). doi:10.1038/nn.4630
- Wadsworth, W. G., Bhatt, H. & Hedgecock, E. M. Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron 16, 35–46 (1996).
- Kennerdell, J. R., Fetter, R. D. & Bargmann, C. I. Wnt-Ror signaling to SIA and SIB neurons directs anterior axon guidance and nerve ring placement in C. elegans. Development 136, 3801–3810 (2009).
- Hedgecock, E. M., Culotti, J. G. & Hall, D. H. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron 4, 61–85 (1990).
- Zallen, J. A., Kirch, S. A. & Bargmann, C. I. Genes required for axon pathfinding and extension in the C. elegans nerve ring. Development 126, 3679–3692 (1999).
- MacNeil, L. T., Hardy, W. R., Pawson, T., Wrana, J. L. & Culotti, J. G. UNC-129 regulates the balance between UNC-40 dependent and independent UNC-5 signaling pathways. Nat. Neurosci. 12, 150–155 (2009).
- Zallen, J. A., Yi, B. A. & Bargmann, C. I. The conserved immunoglobulin superfamily member SAX-3/Robo directs multiple aspects of axon guidance in C. elegans. Cell 92, 217–227 (1998).
- Hutter, H., Wacker, I., Schmid, C. & Hedgecock, E. M. Novel genes controlling ventral cord asymmetry and navigation of pioneer axons in C. elegans. Dev. Biol. 284, 260–272 (2005).
- White, J. G., Southgate, E., Thomson, J. N. & Brenner, S. The Structure of the Nervous System of the Nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 314, 340 (1986).
- Hobert, O. Neurogenesis in the nematode Caenorhabditis elegans. Wormb. online Rev. C elegans Biol. 1–24 (2010).
- Helms, A. W. & Johnson, J. E. Specification of dorsal spinal cord interneurons. Curr. Opin. Neurobiol. 13, 42–49 (2003).
- Thomas M. Jessell. Neuronal Specification in the Spinal Cord: Inductive Signals and Transcriptional Codes. Nat Reviews. 20, 20–29 (2000).
- Kuwada, J. Y. Pioneering and pathfinding by an identified neuron in the embryonic leech. J. Embryol. Exp. Morphol. 86, 155–167 (1985).
- Jacobs, J. R., Goodman, C. S. & Ii, C. N. S. Embryonic Development of Axon Pathways Behavior of Pioneer Growth Cones in the Drosophila. 2412–2422 (1989).
- Kulkarni, R. P., Bak-Maier, M. & Fraser, S. E. Differences in protein mobility between pioneer versus follower growth cones. Proc. Natl. Acad. Sci. U. S. A. 104, 1207–12 (2007).
- Chédotal, A. & Richards, L. J. Wiring the brain: the biology of neuronal guidance. Cold Spring Harb. Perspect. Biol. 2, a001917 (2010).
- Bastiani, M. J. & Goodman, C. S. Guidance of neuronal growth cones in the grasshopper embryo. III. Recognition of specific glial pathways. J. Neurosci. 6, 3542–3551 (1986).
- Hidalgo, A., Urban, J. & Brand, A. H. Targeted ablation of glia disrupts axon tract formation in the Drosophila CNS. 3712, 3703–3712 (1995).
- Hidalgo, A. & Booth, G. E. Glia dictate pioneer axon trajectories in the Drosophila embryonic CNS. Development 127, 393–402 (2000).
- Heiman, M. G. & Shaham, S. DEX-1 and DYF-7 Establish Sensory Dendrite Length by Anchoring Dendritic Tips during Cell Migration. Cell 137, 344–355 (2009).
- Hobert, O. Specification of the nervous system. Wormb. online Rev. C elegans Biol. 1–19 (2005).
- Murray, J. I. et al. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat. Methods 5, 703–709 (2008).
- Murray, J. I. et al. Multidimensional regulation of gene expression in the C . elegans embryo Multidimensional regulation of gene expression in the C . elegans embryo. 1282–1294 (2012). doi:10.1101/gr.131920.111
- Singhal, A. & Shaham, S. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat Commun 8, 1–13 (1AD).
- Norris, C. R. & Kalil, K. Guidance of callosal axons by radial glia in the developing cerebral cortex. J. Neurosci. 11, 3481–3492 (1991).
- Silver, J., Edwards, M. A. & Levitt, P. Immunocytochemical demonstration of early appearing astroglial structures that form boundaries and pathways along axon tracts in the fetal brain. J. Comp. Neurol. 328, 415–436 (1993).
- Minocha, S. et al. Nkx2.1-derived astrocytes and neurons together with Slit2 are indispensable for anterior commissure formation. Nat Commun 6, 6887 (2015).
- Schroeder, N. E. et al. Dauer-Specific Dendrite Arborization in C. elegans Is Regulated by KPC-1/Furin. Curr. Biol. 23, 1527–1535 (2013).
- Ferrario, J. E. et al. Axon guidance in the developing ocular motor system and Duane retraction syndrome depends on Semaphorin signaling via alpha2-chimaerin. Proc. Natl. Acad. Sci. 109, 14669–14674 (2012).
- Wegmeyer, H. et al. EphA4-Dependent Axon Guidance Is Mediated by the RacGAP α2-Chimaerin. Neuron 55, 756–767 (2007).
- Dominici, C. et al. Floor-plate-derived netrin-1 is dispensable for commissural axon guidance. Nature 545, 350–354 (2017).
- Varadarajan, S. G. et al. Netrin1 Produced by Neural Progenitors, Not Floor Plate Cells, Is Required for Axon Guidance in the Spinal Cord. Neuron 94, 790–799.e3 (2017).


 (5 votes)
(5 votes) (No Ratings Yet)
(No Ratings Yet)
 (1 votes)
(1 votes)