Pluripotency in the mouse and beyond…
Posted by Boroviak T, on 4 February 2016
Preimplantation development establishes the founding cell population of the adult mammal in the epiblast. This naïve pluripotent state employs a unique hand of transcription factors to ensure epigenetic resetting and unbiased embryonic potential. In rodents, naïve pluripotency can be captured in the form of embryonic stem (ES) cells1-4, however other mammals have proven more refractory. This raises important questions such as why can naïve pluripotency in rodents be readily stabilised in culture and what is the difference to other mammals, in particular primates, like ourselves? Furthermore, it remains unclear which factors control the rise of naïve pluripotency in the embryo and how primates initiate lineage segregation.
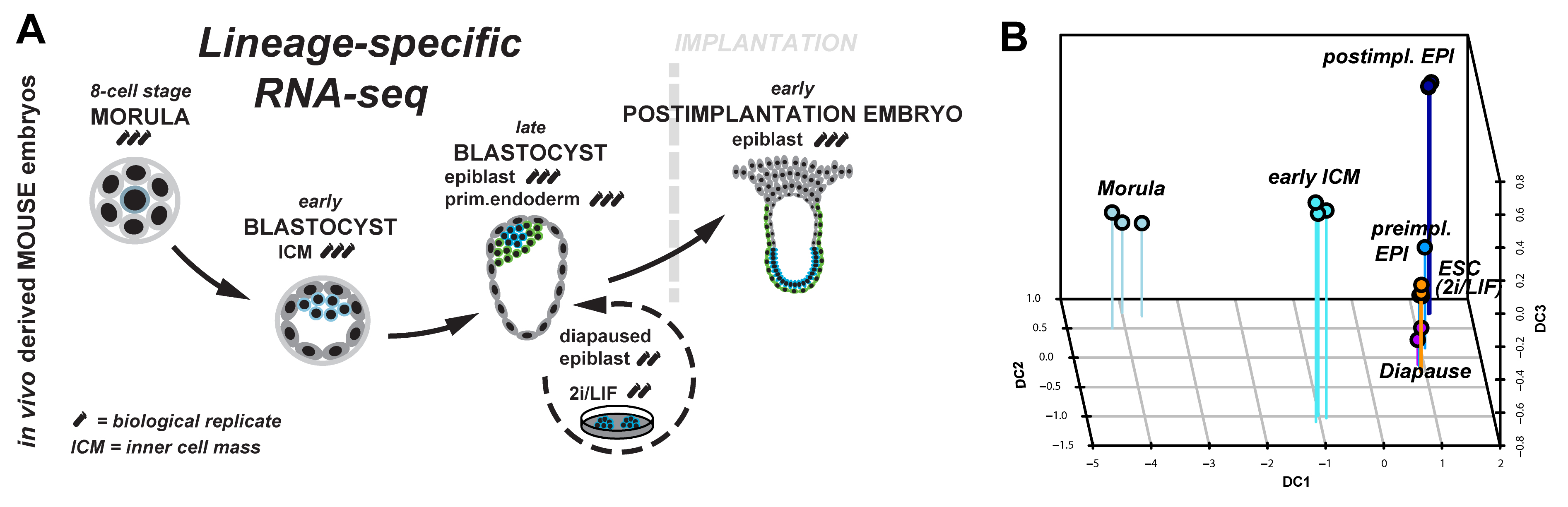
Figure 1: (A) Lineage-specific profiling of the emergence and progression of naïve pluripotency in the mouse embryo. (B) Diffusion map of lineage-specific RNA-seq samples from embryo, diapause and ESC.
We started to tackle some of these questions by scrutinising the emergence and dissolution of naïve pluripotency in vivo5. Taking advantage of Pdgfr::GFP knock-in mice6, we performed lineage-specific RNA-seq from the 8-cell stage to the postimplantation embryo (Figure 1A). This allowed us to define dynamically expressed genes and to derive the transcriptional signature of each developmental stage. Naïve pluripotency markers (Klf2, Klf4, Esrrb, Tfcp2l1) are tightly restricted to preimplantation development, with the majority being expressed in both, embryonic and extraembryonic lineages. Epigenetic modifiers associated with a permissive chromatin state (Tet1, Tet2, Prdm14) were predominant during preimplantation development. These factors were exchanged for regulators mediating de novo DNA methylation (Dnmt1, Dnmt3a, Dnmt3b) and transition to more closed chromatin configuration (Hdac5, Hdac11) after implantation.
Naïve pluripotency is a transient state during normal development. We wanted to relate this to stable pluripotent states in vivo (diapaused epiblasts) and in vitro (ES-cells). Therefore, we profiled diapaused epiblasts and ES-cells using the same protocol. This was essential to allow direct comparison between the minute RNA quantities from the embryo and conventional bulk culture. Analysis of ES-cells and diapause in relation to normal development reveals near-complete conservation of the core transcriptional circuitry operative in the preimplantation epiblast (Figure 1B). Interestingly, diapaused epiblasts showed strong JAK-STAT and WNT signalling pathway component expression. It is tempting to speculate that the pathways evolved to mediate developmental arrest in vivo may facilitate capture of naïve pluripotency in vitro in mouse.
However, what are the differences between naïve pluripotency in the mouse embryo compared to other species, in particular primates? To address this, we turned to the common marmoset (Figure 2), a small New World monkey and emerging primate model for biomedical research7-9. ICMs of early, mid- and late marmoset blastocysts (Figure 3) were isolated by immunosurgery and profiled by RNA-seq. The majority of pluripotency-associated genes was expressed in the ICM and naive markers NANOG, KLF4 and TFCP2L1 colocalised in the putative epiblast. However, the absence of KLF2, NR0B1, FBXO15 and BMP4 suggests primate-specific adaptations in the wider pluripotency network.

Figure 2: Challithrix jacchus (common marmoset )
The specific pathways driving lineage commitment and segregation in the primate embryo are poorly understood, therefore we assessed signalling pathway component expression during normal mouse development, diapause and in the primate ICM. RNA-seq datasets were intersected with annotated signalling pathways for side by side comparison between mouse and marmoset. Ready to use summaries are available to the community through our online resource:
Mouse and Marmoset Pathway Expression Atlas for Early Development
http://pathway-atlas.stemcells.cam.ac.uk/

Figure 3: Immunofluorescence of marmoset late blastocyst for NANOG (green), GATA6 (white), CDX2 (red) and DAPI (blue).
At the early ICM cell stage, we observed substantial differences in NODAL, FGF and WNT signalling. To test the functional relevance, mouse and marmoset morulae were cultured to the late blastocyst stage in the presence of specific inhibitors of these pathways. The experiments demonstrate that lineage specification in primate preimplantation development is regulated by both WNT and FGF pathways, in contrast to mouse, where FGF/ERK is the primary and sufficient driver. Furthermore, the data shows that NANOG expression in the ICM does not require FGF, WNT or NODAL signals. In particular, we noted an increase in NANOG positive cells when ERK activation is inhibited by PD03. Robust expression of Nanog in the absence of FGF/ERK signalling is reported in a variety of species, including mouse, rat, bovine10 and human blastocysts11 and may present a general feature of naïve pluripotency in mammals.
Finally, we would like to emphasise that all lineage-specific RNA-seq data is available as formatted tables in the supplementary to make the data readily accessible for biologists without bioinformatical support.
Literature
1 Evans, M. J. & Kaufman, M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154-156 (1981).
2 Martin, G. R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 78, 7634-7638 (1981).
3 Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519-523, doi:nature06968 [pii] 10.1038/nature06968 (2008).
4 Boroviak, T., Loos, R., Bertone, P., Smith, A. & Nichols, J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol 16, 516-528, doi:10.1038/ncb2965 (2014).
5 Boroviak, T. et al. Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Dev Cell 35, 366-382, doi:10.1016/j.devcel.2015.10.011 (2015).
6 Plusa, B., Piliszek, A., Frankenberg, S., Artus, J. & Hadjantonakis, A. K. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135, 3081-3091, doi:135/18/3081 [pii]
10.1242/dev.021519 (2008).
7 Sasaki, E. et al. Generation of transgenic non-human primates with germline transmission. Nature 459, 523-527, doi:nature08090 [pii]
10.1038/nature08090 (2009).
8 Hanazawa, K. et al. Minimally invasive transabdominal collection of preimplantation embryos from the common marmoset monkey (Callithrix jacchus). Theriogenology 78, 811-816, doi:10.1016/j.theriogenology.2012.03.029 (2012).
9 Mansfield, K. Marmoset models commonly used in biomedical research. Comp Med 53, 383-392 (2003).
10 Kuijk, E. W. et al. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 139, 871-882, doi:10.1242/dev.071688 (2012).
11 Roode, M. et al. Human hypoblast formation is not dependent on FGF signalling. Dev Biol 361, 358-363, doi:10.1016/j.ydbio.2011.10.030 (2012).


 (2 votes)
(2 votes)
 (No Ratings Yet)
(No Ratings Yet)