This interview first featured in Development.
Peter Lawrence, FRS, is a fly geneticist based at the Department of Zoology at the University of Cambridge. During his illustrious career he has carried out pioneering work on pattern formation and polarity, and his contributions have been recognised by many honours, including the Prince of Asturias prize with Gines Morata and election to the Royal Swedish Academy of Sciences. He is also an outspoken critic of the current scientific system and particularly how it affects young scientists. We recently had the opportunity to chat with Peter, and we asked him about the influence of his mentor Sir V. B. Wigglesworth, writing his first grant at age 65 and his time as an editor of Development.

How did you first become interested in biology?
My mum says my first word was ‘butterby’, so I think I was always interested in insects and nature. I started collecting butterflies when I was about 6, using an old tennis racket made into a net. Walter Gehring used to give the impression that his interest in homeotic genes was a gift from very high up, but in my case it was more mundane!
Over the last 20 years you have focused on the problem of polarity, using the fly as a system. Why?
I actually first came across polarity during my PhD, and a small part of my thesis is dedicated to it. I worked on polarity on and off for a decade or so, but then I became more interested in animal design from a genetic point of view. I turned from bugs like Rhodnius to Drosophila in the late 1960s when I came to understand the power of genetics and genetic mosaics. My focus on polarity restarted around 1995, as the result of a chance observation: Jürg Müller and I noticed that bristle orientation was disturbed by Polycomb mutant clones. It reminded me that you can’t build an animal without polarity: vectorial information is absolutely indispensable. The cell has to know not only its position, but also which way to move, to divide, or to build a structure. And yet we know very little about polarity. I thought this was something I could do in my ‘old age’. I had a tenured job at the Laboratory of Molecular Biology (LMB) and Gary Struhl, my collaborator in this project, had a Howard Hughes Medical Institute grant, so we could afford to go on this adventure together. We were both pretty disenchanted with the way science practice was changing, and we wanted to break away from that. We made a policy decision that we would publish the work in Development, because we thought they would take our papers in a fair way, rather than having to worry about ‘appearances’. We are still collaborating and discussing a new paper even now, which will be destined for Development. We work with genetic mosaics because polarity is a contextual problem, not a single cell problem. If you say that a bristle points towards the back of the fly, then the cell has to know where the back of the fly is, and has to compare itself with its neighbours. So mosaics are a really valuable tool.
You did your PhD with the famous insect physiologist Sir V. B. Wigglesworth. How did this time influence your career?
He influenced me a lot. Wigglesworth was an unusual man. He let us get on with things and didn’t give us much direction. But I think his idea that you should be free to do what you want is rather important. You struggle perhaps, but you learn, and when you discover something it is very rewarding because you haven’t been told to discover it, you haven’t been told what to find or which ideas are in fashion. I remember Wigglesworth telling me early on, when I was reading some obscure paper in German: ‘Lawrence, you shouldn’t be reading too much. You should get on with looking at things yourself.’ It is very important to realise that if you want to do something new it is better to look at the material; if you want to do what everybody else is doing then it is better to listen to them. He also didn’t put his name on any of our papers. He wrote around 264 papers, of which all but 19 were single-author papers. And he kept on doing research until he was about 90. Wigglesworth’s example and ideas have affected my whole life.
You are also known for not adding your name to a paper from your lab unless you feel that you have made a significant contribution. Why do you do this and would you recommend this practice to someone setting up their own lab now?
The issue of paper authorship is a question of scientific integrity. If you aren’t responsible for what is in the paper, because you don’t know what has been done or you can’t assess the quality of the work therein, and you haven’t done it anyway, why should your name be there? I would feel very uncomfortable if I wasn’t sure whether the work that was in the paper was honest and correct. There is also the question of stealing credit. Nowadays people very often don’t get credit for what they do. When they are young they don’t get the credit, and when they are older they get the credit for what other people do. It is not logical, scientific or honest, so I would like to resist it – even more than I have been able to do, especially lately.
The truth is that in the present circumstances it can be imprudent not to put your name on the work of your students and postdocs, because measurement has almost destroyed the possibility of being honest. If you don’t put your name on things then you won’t get credited for it. If you don’t get the credit, you won’t get a tick on measures like your H factor or your citation index, which count so much in getting grants and positions. It is very sad, but true, that I wouldn’t advise a young person to go against present-day practices. Later, when they become established, they should try to change the system by political action and also by establishing different policies in their own lab.
You wrote your first grant at age 65, when you left the LMB. What was it like and what did it tell you about the current state of science?
Leaving the LMB after so many years was traumatic, particularly as I felt I was subjected to age discrimination. I had to find somewhere to work so I approached the Departments of Zoology and Genetics at the University of Cambridge. They both said that they wanted me and I thought, ‘That’s nice, they must remember me from the old days!’ I was very naïve as I hadn’t understood the importance of the REF (Research Excellence Framework; now the RAE, Research Assessment Exercise): my coming to the Zoology Department (which was the one I chose) with a pile of papers from the last few years would give them a significant financial benefit. There are a lot of issues surrounding this system that concern me, but I was very glad to be accepted by the Zoology Department, who have been good to me.
The department would take me but I needed a grant. The LMB is core funded, so I never had to write a grant before. I actually went on an MRC grant-writing course, which was quite amusing because everyone else there was 35 years younger and at the end of their postdocs. I think the application I wrote was honest, and when I showed it to some helpful colleagues, they all came back with the same message: ‘You can’t write this. You can’t say the truth and you can’t indicate that your experiments may not work or that there may be some doubt as to the validity of your approach.’ With that lesson I went back and wrote something that felt more and more fictionalised. I must have learned sufficiently though, because I have written four main grants now and the Wellcome Trust has kindly funded all of them. The process is a bit useful as it can force you to think about what you might do, but what you actually write down is not usually what you do in practice. And if you have a really good idea you don’t put it in the grant, because someone might steal it.
Over the years, I have come to realise that grant application is like a game. You have to follow the rules and the people who figure out how to play the game successfully do well. We write plausible, feasible grant applications as evidence that we have reasonable ideas and know what we are doing. The scientists reading them look for technical validity, intellectual coherence and a proper purpose. Yet we all know that the chance you will actually do the experiments is remote. It’s a game. But it is an expensive game in terms of time and emotional energy.
How could it be made less wasteful?
The Wellcome Trust has been trying to do this and they have made big improvements, primarily by reducing the length of grant applications and reintroducing interviews. But there is still a huge amount to do. Remember that in Europe four grant applications have to be written for every one that is approved! PIs spend so much of their time writing grant applications, most of which are going to fail. The grant-giving organisations spend most of their time assessing grant applications that they are going to fail. I think the grant application process could be improved by becoming more realistic and much more streamlined.
Another thing that would simplify things would be to change the emphasis away from what people pretend to want to do towards assessing what they have done. We could ask: ‘Has this person done anything any good in the last 3-4 years?’ If so, let’s give them another 3-4 years. If they haven’t, let’s give them an option. They either write a new application, which we will assess in the old way, or they give up and we don’t fund them. That would save a lot of time.
In the late 1990s, you started writing a series of articles expressing your views on many issues of the current scientific system. How do you think the situation has changed since you wrote these articles?
In 1996, I was invited to give the inaugural Wigglesworth lecture at the XX International Congress of Entomology in Florence. I talked mostly about his work, but at the end I described his approach to science, including what we have discussed above. I never had such a response to a lecture before. I realised that there was a great need for somebody to tell the truth about what was happening to science. Particularly for young scientists and their prospects: how they are being exploited and manipulated, and how their initial dreams of science as an exciting pursuit are being despoiled. So I started writing a series of articles, and they are read much more than my research papers.
I have realised that the most fundamental evil is metrics, the idea that people can be measured. Once you introduce this concept then everybody tries to meet the measurement. This is true throughout society, not just in science. But it has destroyed the heart of science, spoiling what we actually write and mitigating against originality. And things have gotten much worse since I started writing these articles. There is a huge momentum, and it is partially fuelled by the fact that many scientists train too many young people. This creates too many people applying for relatively few jobs. The response has been to make even more phony measurements to discriminate between them. Publishing in top journals has become so important to get jobs and this is particularly problematic. Everyone now believes that you can’t get far without a good paper in one of these top journals. This is partially true, but it is a myth that poisons the system.
I think there is more awareness of these problems now, but I’m not convinced the people with the most power are keen to change the system – because they benefit from it. Getting your paper in a top journal is a skill, a bit like the game of grant writing. It is the skill of manipulating your paper to make it attractive to the editors of those journals and choosing the topic according to current fashion. And this is why I think the current system mitigates against originality. In short, I don’t think it has got better, I think it has got worse.
Over the years, many members of your lab have gone on to establish successful labs. Is mentoring something that is important for you?
Very important. I actually had very few people in my lab, and maybe that’s why I was able to give them more time. Generally speaking I only ever had one graduate student at a time, so over 53 years, fewer than 15 students. I also never had more than one or two postdocs at once. My theory is also that by giving them freedom, they like science because they find it rewarding. So all of them developed a taste for discovering things, and I am very proud of that.
Would you enforce lab size limits if you could?
Yes. Grant-giving bodies have a limited pot of money, so why give so much to a small number of very large labs? Why not look at the efficiency of those top labs per person? There was a study that worked out the most productive group size (in terms of publications per person). The answer came out at about 6.5, which means that all those labs that have group sizes above that, 10-20 lab members being quite common, are on average more inefficient per person. They attract lots of students because students are misled by the same myth. If a group produces a Cell paper once every two years, the student applying thinks, ‘I’m going to get a Cell paper’. But maybe there are 20 people in the group, and only one of them has been involved in that Cell paper. What happens to the other 19? So I would ask grant-giving bodies not to keep feeding successful scientists with more and more money and more and more students.
You are known for encouraging younger scientists to think about the bigger picture in their research. Do you think this kind of thinking is lacking?
It has always been lacking. Max Perutz specifically advised young people to think of a big, unsolved problem that guides you like a lighthouse when you make decisions. Do these experiments give me a chance of moving towards an understanding of this big problem? Of all the scientists I have got to know well, the one I admire the most is Francis Crick. He was an extreme case of ‘bigger picture’ thinking. His first choice of problem was the difference between the living and the non-living. It is such an obvious thing, but people hadn’t thought of it as a scientific problem that should be addressed. Everyone should try this approach. Maybe not in such a grand way, but they should think about what for them is an interesting mystery that needs a solution, and work towards it. You don’t get to it straight away, but you need it out there as a guiding light.
You were an editor with Development for over 30 years. How was your experience at the journal, and how did the field (and the journal) change during this period?
When I started as an editor on the journal it was still called JEEM (Journal of Embryology and Experimental Morphology). It was a very old-fashioned journal, limited in scope and not concerned with being trendy. Then, in 1987, Development started, as an attempt to modernise the whole business, both technically and scientifically. We had a group of editors keen to make Development a more successful journal. I think we did a good job and it was fun to be part of that process.
During this period, something else happened that affected Development. The purpose of publishing was recast, from producing papers of scientific record that stimulated and educated others, to instead getting tokens necessary for survival in the scientific system. That recasting had a huge effect on the publishing process and Development was not immune. Other competitive journals appeared. For example, when Developmental Cell came into existence we thought we could compete with it on the grounds of quality. And we could really, but Developmental Cell had the Cell marketing logo on it, so people were tempted to publish there instead. I think the recent changes in the publishing world have done considerable damage to journals like Development. Of course it still has a great reputation, and if you look at the citation lifetime of Development papers you will see that it is very long because the journal publishes papers that have internal quality without necessarily the pizzazz needed for other journals. But it is perhaps not as well respected as it deserves to be. I don’t think there is anything we could have done about it, but quality wasn’t enough.
But I enjoyed being an editor of Development; I thought it gave me a chance to help people who sometimes were being badly treated by the system, to give them a chance to publish. I also tended not to be as impressed by fashion as other editors, particularly younger ones in other journals, might have been. Having been around for so long, I could see how trends come and go.
What would people be surprised to find out about you?
I really enjoy the theatre, so we go nearly every week to London to watch plays. I am also a mad keen gardener. We don’t have a television, which I think is quite unusual, mostly because we have too many other things to do.
 (8 votes)
(8 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)
 (8 votes)
(8 votes)



 Peter Lawrence, FRS, is a fly geneticist based at the Department of Zoology at the University of Cambridge. During his illustrious career he has carried out pioneering work on pattern formation and polarity, and his contributions have been recognised by many honours. He is also an outspoken critic of the current scientific system and particularly how it affects young scientists. We recently had the opportunity to chat with Peter, and we asked him about the influence of his mentor Sir V. B. Wigglesworth, writing his first grant at age 65 and his time as an editor of Development. See the Spotlight article on p.
Peter Lawrence, FRS, is a fly geneticist based at the Department of Zoology at the University of Cambridge. During his illustrious career he has carried out pioneering work on pattern formation and polarity, and his contributions have been recognised by many honours. He is also an outspoken critic of the current scientific system and particularly how it affects young scientists. We recently had the opportunity to chat with Peter, and we asked him about the influence of his mentor Sir V. B. Wigglesworth, writing his first grant at age 65 and his time as an editor of Development. See the Spotlight article on p.  The development, homeostasis and regeneration of tissues result from a complex combination of genetics and mechanics. Sugimora, Lenne and Graner describe techniques to measure forces in cells and tissues, and discuss their applications in developmental contexts. See the Primer on p.
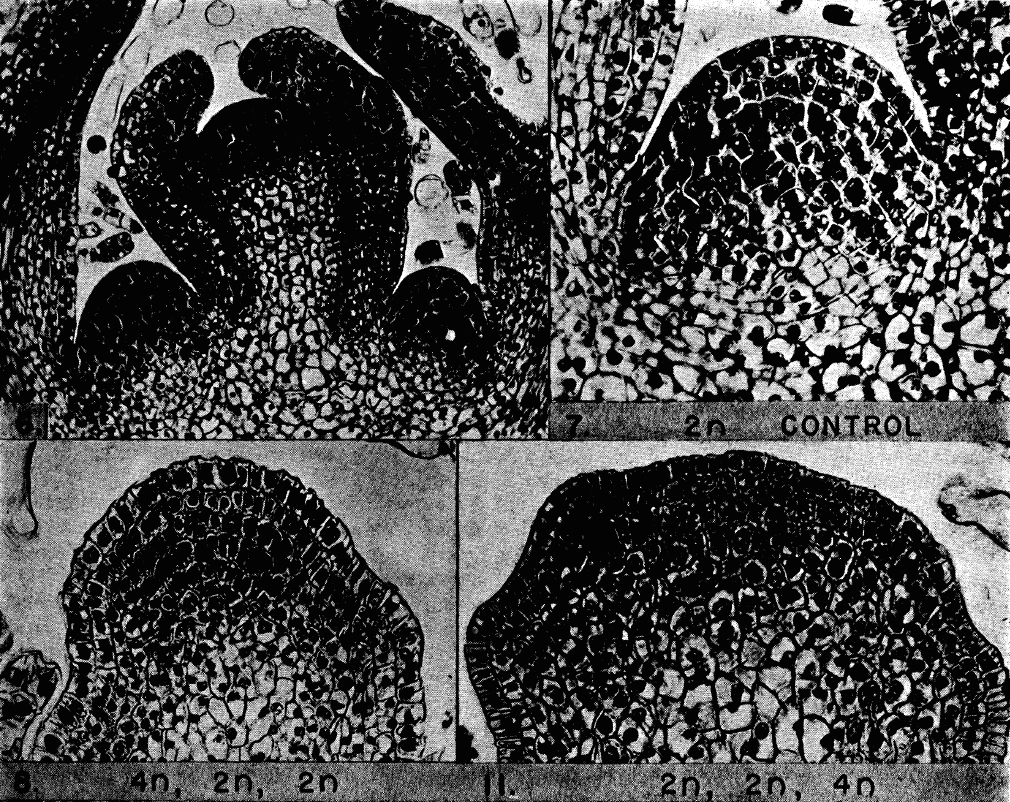
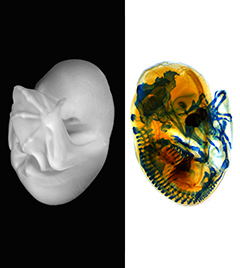
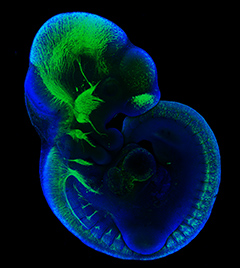
The development, homeostasis and regeneration of tissues result from a complex combination of genetics and mechanics. Sugimora, Lenne and Graner describe techniques to measure forces in cells and tissues, and discuss their applications in developmental contexts. See the Primer on p. 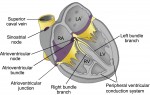 The cardiac conduction system (CCS) consists of distinctive components that initiate and conduct the electrical impulse required for the coordinated contraction of the cardiac chambers. Here,
The cardiac conduction system (CCS) consists of distinctive components that initiate and conduct the electrical impulse required for the coordinated contraction of the cardiac chambers. Here, 
 (17 votes)
(17 votes)

 Marcos and Julia were a brilliant young couple who instantly made the laboratory smart and fun. Marcos took up cancer research, the second person to ever work on this disease in the laboratory. He loved everything about conducting science and he was ambitious. He worked crazy hours. With a laconic Argentinian accent, he kept up a running science conversation that lasted until now. Julia worked on separate topics and they lived balanced lives. Marcos loved board sports from snowboarding to kite- and wind-surfing and was superb at them. Marcos and Julia made friends easily. The Facebook postings and the emails we’ve received and the conversations we’ve had over the past few days are a reminder just how much they were loved. We all rooted for their success because they were really good people.
Marcos and Julia were a brilliant young couple who instantly made the laboratory smart and fun. Marcos took up cancer research, the second person to ever work on this disease in the laboratory. He loved everything about conducting science and he was ambitious. He worked crazy hours. With a laconic Argentinian accent, he kept up a running science conversation that lasted until now. Julia worked on separate topics and they lived balanced lives. Marcos loved board sports from snowboarding to kite- and wind-surfing and was superb at them. Marcos and Julia made friends easily. The Facebook postings and the emails we’ve received and the conversations we’ve had over the past few days are a reminder just how much they were loved. We all rooted for their success because they were really good people.