Nature’s most interesting secrets can sometimes be found in our own backyards. One such secret is related to all birds, those pigeons, thrushes and sparrows that we see everyday. This familiarity means that we do not think too much of birds passing them by on our way to work or school. However, if the birds no longer existed, wiped out by an ancient cataclysm, and were only known to us through their fossil remains, we would be truly awed by them, by their strange anatomy, by how different they would appear from all other vertebrates, including their own ancestors. Modern birds are the remaining descendants of once glorious lineage of theropod dinosaurs, the bipedal carnivorous reptiles who dominated the landscape during the Mesozoic Era. The theropods had long and powerful snouts with many teeth, relatively small eyes and brains – a far cry from modern birds that have no teeth, their skulls featuring relatively large eyes and brains. Instead of a face with a snout constructed from many bones, birds have an elongated bill, composed largely of just two bones – one bone of the upper beak (premaxillary bone) and one for the lower jaw (mandibular bone). The beaked face of the modern bird looks distinctly different from the faces of their ancestors. How and why the birds acquired their unique craniofacial appearance?
In 2012 my group published Bhullar et al., 2012 report where we explained the first part of the story of the avian skull and face [1]. We studied skulls of the modern and primitive birds, non-avian theropod dinosaurs and other members of the Archosauria, a group of amniotes, which originated about 250 million years ago and included crocodilians, pterosaurs, all dinosaurs and birds. Landmarks were placed on each skull at homologous positions that allowed us to compare archosaurian skull shapes as they evolved and changed over millennia. We used computer software-based geometric morphometric and principal component (PCA) analyses (Fig.1). To better understand the role of developmental changes in this evolutionary story, we also included late embryos and juveniles of both birds and dinosaurs, where available. First, we noticed that the skulls of adult theropods became longer as they evolved from the more basal archosaurs. Second, the skulls of the theropod embryos and juveniles were morphologically different from adults occupying a distinct area of morphospace – they had shorter faces, fewer teeth and proportionally larger brains and eyes. Lastly, the first birds, such as the famed Archeopteryx and Confuciusornis were found precisely in the theropod “baby space”. Ontogenetically, the early birds appear to undergo little change from juvenile to adult states. For example, the skull of the young Eichstätt specimen of Archaeopteryx is about half size of the Berlin specimen’s skull but has a nearly identical cranial morphology, and, importantly, both of them resemble the juvenile theropod skulls. Thus, we concluded that birds are paedomorphic in terms of their cranial morphology in retaining a morphology as adults that resembles that of the juveniles or embryos of their ancestors. Birds are dinosaurs, whose heads and faces never grow old…
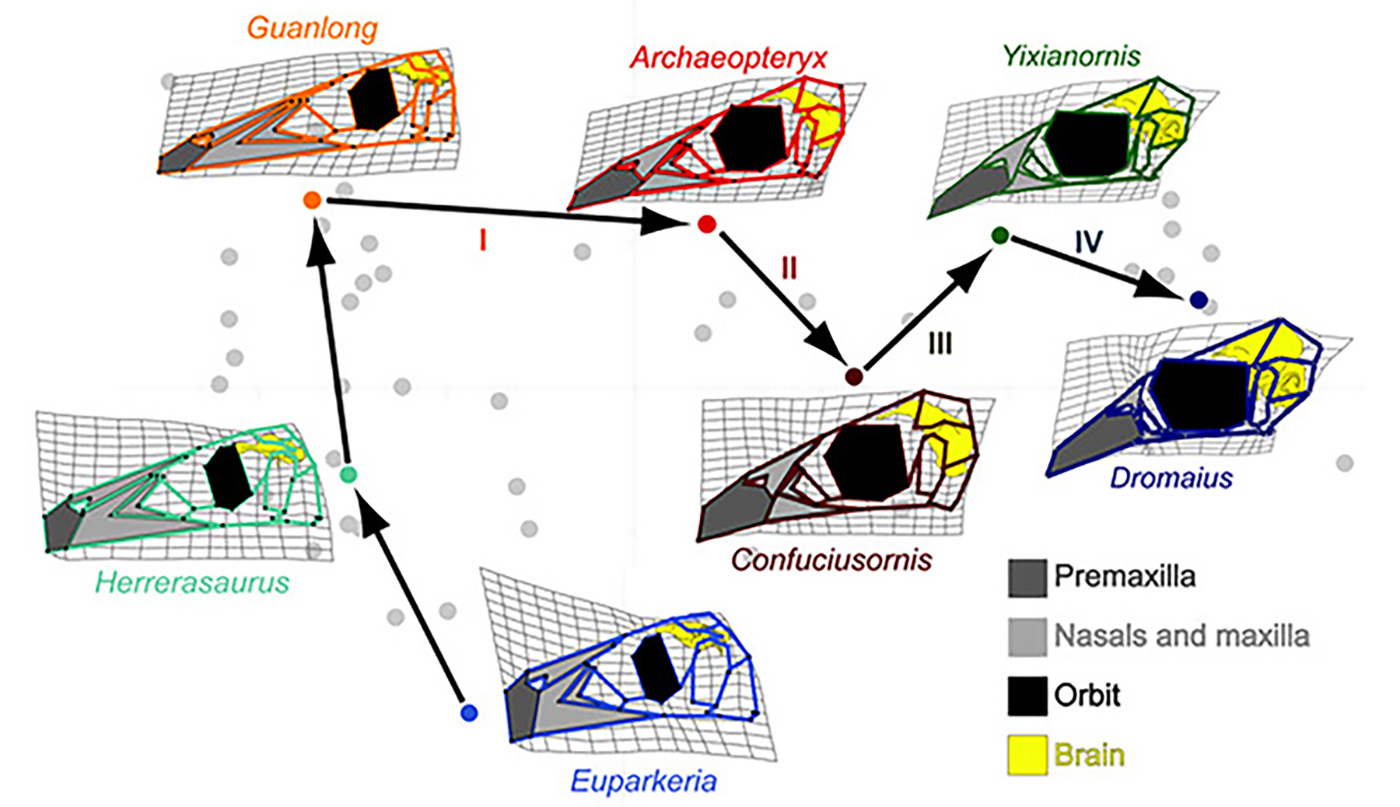
Figure 1. A complicated trajectory of skull shape changes during evolution of non-avian archosaurs and birds. Note the collapse of the face (light grey) and subsequent expansion of the beak 9dark grey) in birds. Euparkeria is an early archosaur, Herrerasaurus is one of the one of the earliest dinosaurs. Guanlong is Guanlong is a tyrannosauroid theropod dinosaur. Archeopteryx is the first bird. Confuciusornis is another early bird. Yixianornis is a beaked bird from the early Cretaceous period. Dromaius (emu) is a basal modern bird [1].
Paedomorphosis is an example of heterochrony, a change in timing or order of developmental events that produces a phenotypic alteration in the adult. There are two ways by which paedomorphosis can arise [2]. One is called neoteny (juvenilization), when somatic development of an animal is slowed or delayed so at the onset of sexual maturity it still retains juvenile characteristics. One of the best-known examples of neoteny is certain species of urodele amphibians, which sexually mature despite preserving a number of larval features, such as a finned tail and external gills. The second type of paedomorphosis is called progenesis when sexual development occurs faster than in the ancestral lineage and results in early somatic maturation and juvenile-looking adults. Paedomorphosis in the bird lineage is probably one of the most striking examples of progenesis reported to date. Our PCA graph shows that birds have significantly shorter ontogenetic trajectories than in all other studied archosaurs. Importantly, these short trajectories also match histological data indicating that sexual and somatic maturation times were truncated during the paedomorphic transformation. So, developmental program in birds is much shorter because they become mature much faster (up to 20 times faster!) than more basal archosaurs.
There was a point in early avian evolution when the shapes of their skulls were nearly perfectly pedomorphic. The modern birds, however, look substantially different from those first birds. Archeopteryx, famously, still had a snout with teeth and, while baby-like, it still looked like a dinosaur with wings. Modern birds have a beak and our latest publication sheds light on how this unique structure evolved from a snout [6]. Modern birds number about 10,000 species, the largest and arguable most successful group of land vertebrates [3]. Much of their success can be attributed to variation in beak shape. Birds use beaks of a bewildering variety of different designs to obtain food items (hooked beaks of predatory birds, long and straight beaks of herons and hummingbirds, deep beaks of finches and sparrows, serrated beaks of mergansers and flamingos for catching fish and filter feeding, respectively), to build or weave nests (most birds), to dig burrows in the ground (puffins and swallows), to hammer or excavate wood to build a hollow enclosure (woodpeckers and chickadees), to dissipate heat (toucans) and to communicate (song birds, storks, owls). Above all, the bird beak is a precision grasping tool, and its exact shape is often critical to its function. In fact, it is likely that bird beaks evolved to replace the long and agile fingers of their theropod predecessors. It is also a great example of a key evolutionary innovation, a novelty that had dramatic phylogenetic consequences [4]. But how the bird beaks originate?
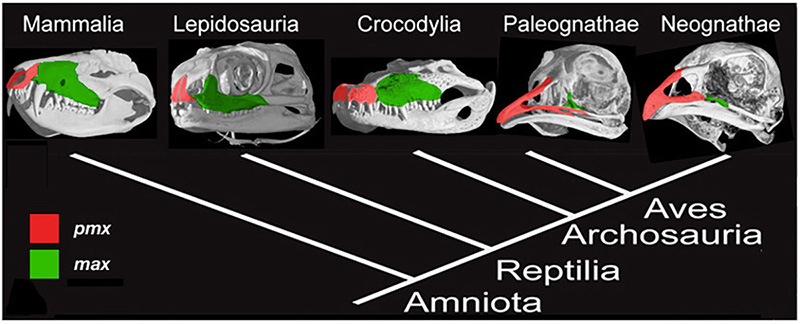
Figure 2. Amniote phylogeny with skull μCT scans and configuration of the premaxillary (red) and maxillary (green) bones. Mammals (Mammalia), lizards (Lepidosauria), and alligators (Crocodylia) all have paired, small and rounded premaxillary bones. Modern birds have fused, large and elongate premaxillae [6].
The upper beak in birds is a particular challenge to understand. It is a novel structure, which forms as a result of rostral expansion of the fused premaxillary bone, which grows as two small capping elements in basal archosaurs and most dinosaurs but expands to structurally and functionally dominate the upper face in modern bird lineage (Fig.2). The remainder of the snout and much of the face in birds are paedomorphically truncated. To address the origin of the avian beak, my group implemented the following research program, developed for studying developmental mechanisms for evolutionary novelties [5]: first, we traced the transformation of the distinctive avian facial skeleton in the fossil record using geometric morphometrics; then we examined candidate gene expression domains in reptiles and birds to phylogenetically polarize expression patterns, and, lastly, experimentally restored the inferred ancestral craniofacial expression patterns in developing chicken embryos [6]. To analyze the results of functional experiments, we included the resultant cranial phenotypes in the broader geometric morphometric analysis, which contained ancestral snouted and bird-faced fossil forms.
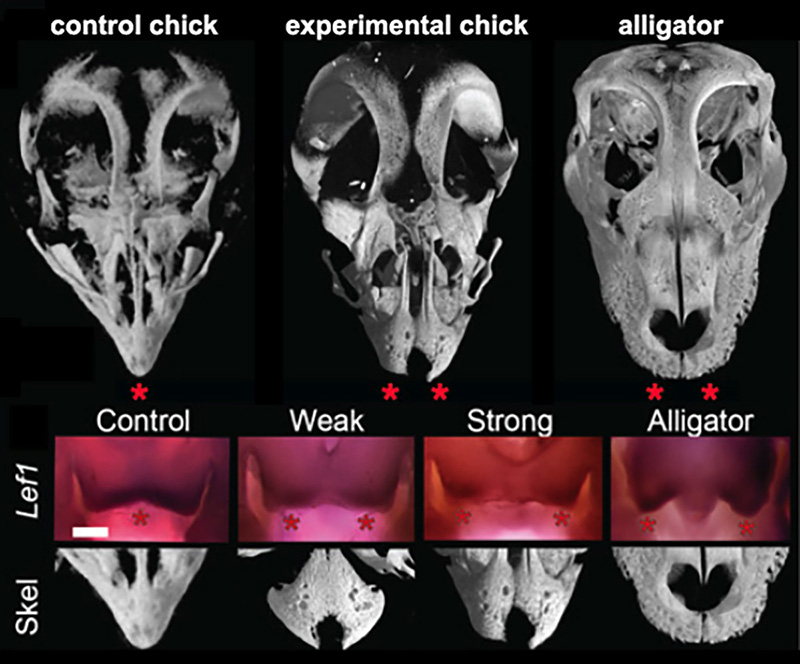
Figure 3. Results if functional experiments reveal roles of FGF and WNT signaling in the origin of modern bird beak. μCT scans of alligator skulls, wild- type chick, and FGF inhibitor-treated chick embryonic skulls. Note the large and paired pmx in the inhibitor-treated embryo (stars). Suppression of FGF signaling causes a range of phenotypes including a suppression of medial WNT signaling and completely separated and round pmx bones [6].
The principal-component analysis of shape variation in premaxilla of a range of extant and extinct archosaurs (including embryos, juveniles, and adults) revealed that adult and embryonic modern birds were separated from the ancestral forms along in having relatively long, narrow premaxillae and straight-edged premaxillae. Predictably, premaxillae of the first bird Archeopteryx clusters with the non-avian snouted dinosaurs. Next, we determined that the early medial expression of Fgf8 and later medial WNT signaling in the developing faces is phylogenetically exclusive to birds. These molecules are expressed in the right time and the right place to control the formation of snout and beak. Embryos of all other amniotes studied (mouse, turtle, lizard, alligator) had only two lateral expression domains around the nasal pits. We then placed silica bids soaked with either FGF or WNT specific inhibitors to replicate an inferred ancestral pattern. Near-hatching skeletal phenxotypes of both FGF-inhibited and WNT-inhibited experimental embryos were not simply abnormal – they were similar to each other and strikingly resembled ancestral archosaurian forms in having abbreviated, rounded, and partially or fully paired premaxillae (Fig.3). Embryos with strong phenotypes displayed complete separation of the premaxillae with a suture-like division between them. We did not find any evidence of cell death playing a role in generating this atavistic phenotype and instead found evidence for a loss of the fusion zone between the two premaxillae. Interestingly, In addition to the premaxillary bone phenotype, the palates of experimental embryos also showed an altered and more ancestral phenotype. The palatine bones of modern birds have a distinctive shape that allows for a kinetic movement of the upper beak relative to the skull while it fuses broadly with the facial bones and cranial base in other archosaurs.
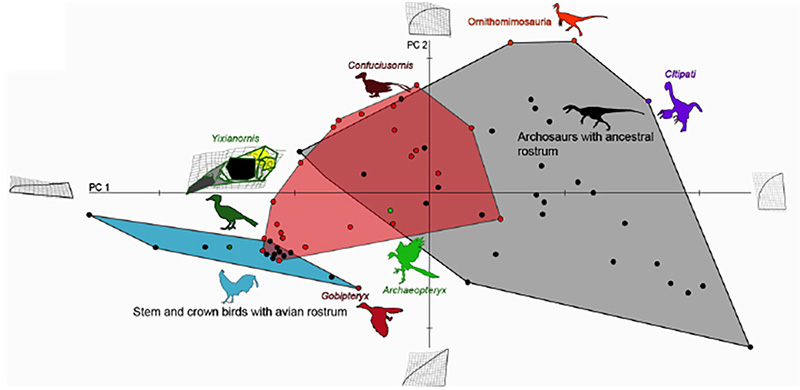
Figure 4. Comparative analysis of premaxillary shapes in experimental embryos with those from modern birds, primitive birds and non-avian archosaurs. Control embryos and experimental embryos showing near normal phenotypes clustered with forms possessing an avian beak (blue polygon), all of the embryos with the more pronounced phenotypes (red polygon) fell outside of the cluster of beaked birds and most of them instead clustered with more basal archosaurs (grey polygon), suggesting that the ancestral bone shapes were indeed successfully resurrected [6].
This study offers an example how we can probe the intrinsic mechanisms for even large-scale evolutionary transformations. The precise genetic/genomic nature of the regulatory change in expression of Fgf8 remains to be investigated but its implications for both development and evolution of birds are clear and profound. Other molecules and pathways are likely to be involved to explain the full scope o the transformation of the reptilian skull and snout into an avian face and beak. Curiously, we have shown that while the fossil record can provide useful developmental hypotheses, the intermediate phenotypes resulting from our experimental restoration of the inferred ancestral developmental patterns in return predict undiscovered transitional morphologies in the fossil record, yet undiscovered avian ancestors with gap-bridging facial bone phenotypes (Fig.4). It may also be significant that two of the most important modern bird beak bones (pmx and palatine) are regulated by the same pathways and are, thus, integrated developmentally and evolutionarily.
References:
Bhullar, B., Marugán-Lobón, J., Racimo, F., Bever, G., Rowe, T., Norell, M., & Abzhanov, A. (2012). Birds have paedomorphic dinosaur skulls Nature, 487 (7406), 223-226 DOI: 10.1038/nature11146
Alberch,P., Gould,S.J., Oster,G.F., & Wake,D.B. (1979). Size and shape in ontogeny and phylogeny Paleobiology , 5, 296-317
Gill, F. B. Ornithology. Freeman, 2006
Hodges, S., & Arnold, M. (1995). Spurring Plant Diversification: Are Floral Nectar Spurs a Key Innovation? Proceedings of the Royal Society B: Biological Sciences, 262 (1365), 343-348 DOI: 10.1098/rspb.1995.0215
Mallarino, R., & Abzhanov, A. (2012). Paths Less Traveled: Evo-Devo Approaches to Investigating Animal Morphological Evolution Annual Review of Cell and Developmental Biology, 28 (1), 743-763 DOI: 10.1146/annurev-cellbio-101011-155732
Bhullar, B., Morris, Z., Sefton, E., Tok, A., Tokita, M., Namkoong, B., Camacho, J., Burnham, D., & Abzhanov, A. (2015). A molecular mechanism for the origin of a key evolutionary innovation, the bird beak and palate, revealed by an integrative approach to major transitions in vertebrate history Evolution DOI: 10.1111/evo.12684
 (10 votes)
(10 votes)
 Loading...
Loading...
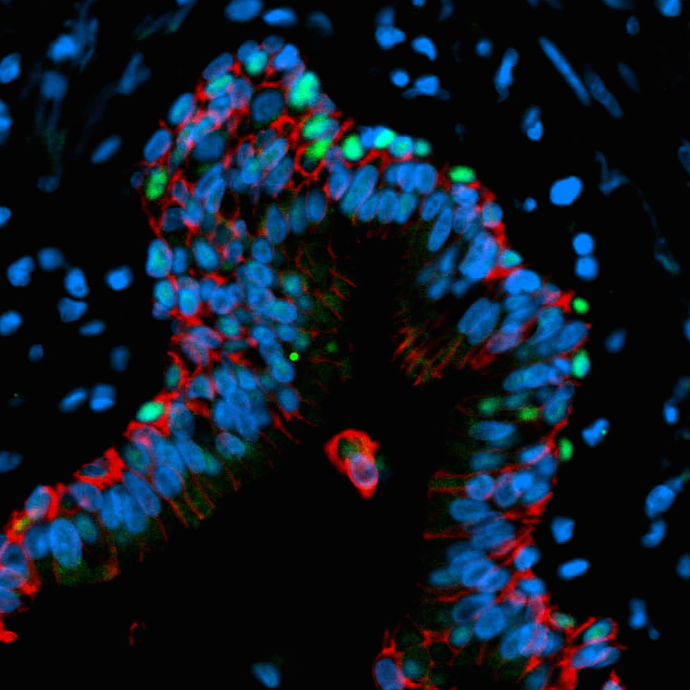 – Briana discussed how she and her colleagues generated human lung organoids in vitro, work published in eLife.
– Briana discussed how she and her colleagues generated human lung organoids in vitro, work published in eLife.




 (No Ratings Yet)
(No Ratings Yet)



 (3 votes)
(3 votes)








 (10 votes)
(10 votes)

 The Hippo signalling pathway regulates organ growth: activation of the pathway inhibits proliferation and promotes apoptosis. The core pathway consists of two kinases, Hippo and Warts, along with their adaptor proteins. Warts phosphorylates and thus inactivates the transcriptional co-activator Yorkie, whose target genes include regulators of cell proliferation and survival. Upstream of Warts, various factors impact on kinase activity, including cytoskeletal and polarity regulators. While Hippo signalling is best known for regulating growth, on p.
The Hippo signalling pathway regulates organ growth: activation of the pathway inhibits proliferation and promotes apoptosis. The core pathway consists of two kinases, Hippo and Warts, along with their adaptor proteins. Warts phosphorylates and thus inactivates the transcriptional co-activator Yorkie, whose target genes include regulators of cell proliferation and survival. Upstream of Warts, various factors impact on kinase activity, including cytoskeletal and polarity regulators. While Hippo signalling is best known for regulating growth, on p.  The hair cells (HCs) of the inner ear mediate both our auditory and our vestibular senses. Following loss or damage, mammalian HCs show very limited capacity to regenerate, creating a therapeutic need to generate new HCs in vitro for cell replacement strategies. Domingos Henrique and co-workers now report a relatively simple and efficient protocol for deriving HCs from mouse embryonic stem cells, by expression of key transcription factors involved in HC differentiation during development (p.
The hair cells (HCs) of the inner ear mediate both our auditory and our vestibular senses. Following loss or damage, mammalian HCs show very limited capacity to regenerate, creating a therapeutic need to generate new HCs in vitro for cell replacement strategies. Domingos Henrique and co-workers now report a relatively simple and efficient protocol for deriving HCs from mouse embryonic stem cells, by expression of key transcription factors involved in HC differentiation during development (p. 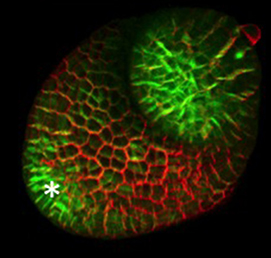 The arrangement of leaves and flowers around the stem typically follows a stereotyped phyllotactic pattern, with the location of a new leaf defined by the positions of previously specified ones. This is controlled by the plant hormone auxin. In the meristem epidermis, auxin accumulates at the site of future primordia, generating intervening regions of auxin minima where no new organ can form. Auxin-dependent phyllotactic patterns can be computationally modelled in two dimensions, considering only the epidermal layer. However, auxin flows into lower layers of the primordium, where the incipient midvein is thought to drain auxin away from the meristem. The contribution of this auxin drainage to leaf development has been hard to assess, but Didier Reinhardt and co-workers (p.
The arrangement of leaves and flowers around the stem typically follows a stereotyped phyllotactic pattern, with the location of a new leaf defined by the positions of previously specified ones. This is controlled by the plant hormone auxin. In the meristem epidermis, auxin accumulates at the site of future primordia, generating intervening regions of auxin minima where no new organ can form. Auxin-dependent phyllotactic patterns can be computationally modelled in two dimensions, considering only the epidermal layer. However, auxin flows into lower layers of the primordium, where the incipient midvein is thought to drain auxin away from the meristem. The contribution of this auxin drainage to leaf development has been hard to assess, but Didier Reinhardt and co-workers (p.  The eggs of drosophilid species are characterised by the presence of dorsal appendages, which arise from the follicular epithelium. Different species display different numbers and morphologies of appendages, but little is known about how these differences arise from a morphological perspective. Now, on p.
The eggs of drosophilid species are characterised by the presence of dorsal appendages, which arise from the follicular epithelium. Different species display different numbers and morphologies of appendages, but little is known about how these differences arise from a morphological perspective. Now, on p. 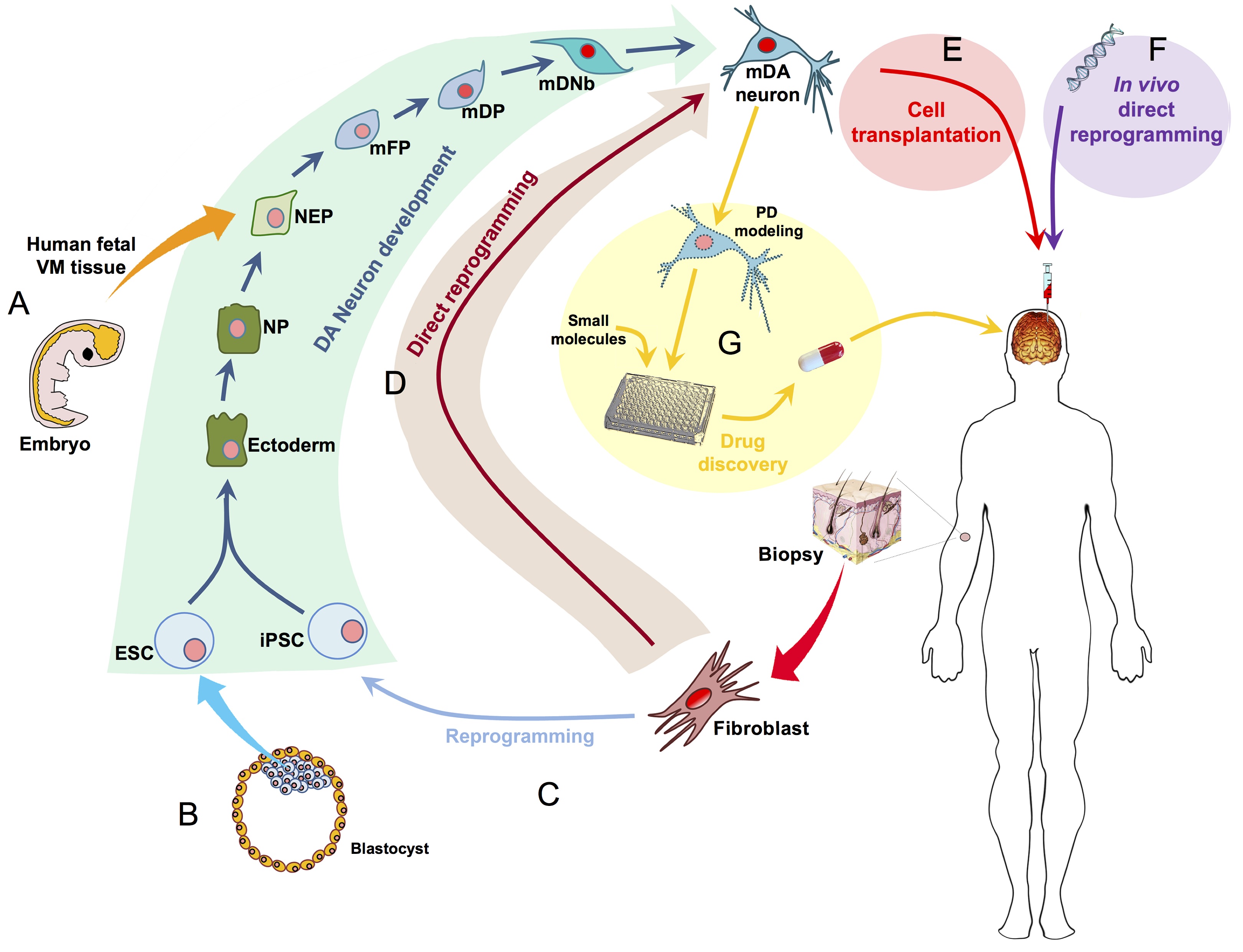 Ernest Arenas, Mark Denham and Carlos Villaescusa summarise recent efforts to generate human midbrain dopaminergic neurons in vitro, from pluripotent stem cells or from somatic cells via direct reprogramming. See this Primer on p.
Ernest Arenas, Mark Denham and Carlos Villaescusa summarise recent efforts to generate human midbrain dopaminergic neurons in vitro, from pluripotent stem cells or from somatic cells via direct reprogramming. See this Primer on p. 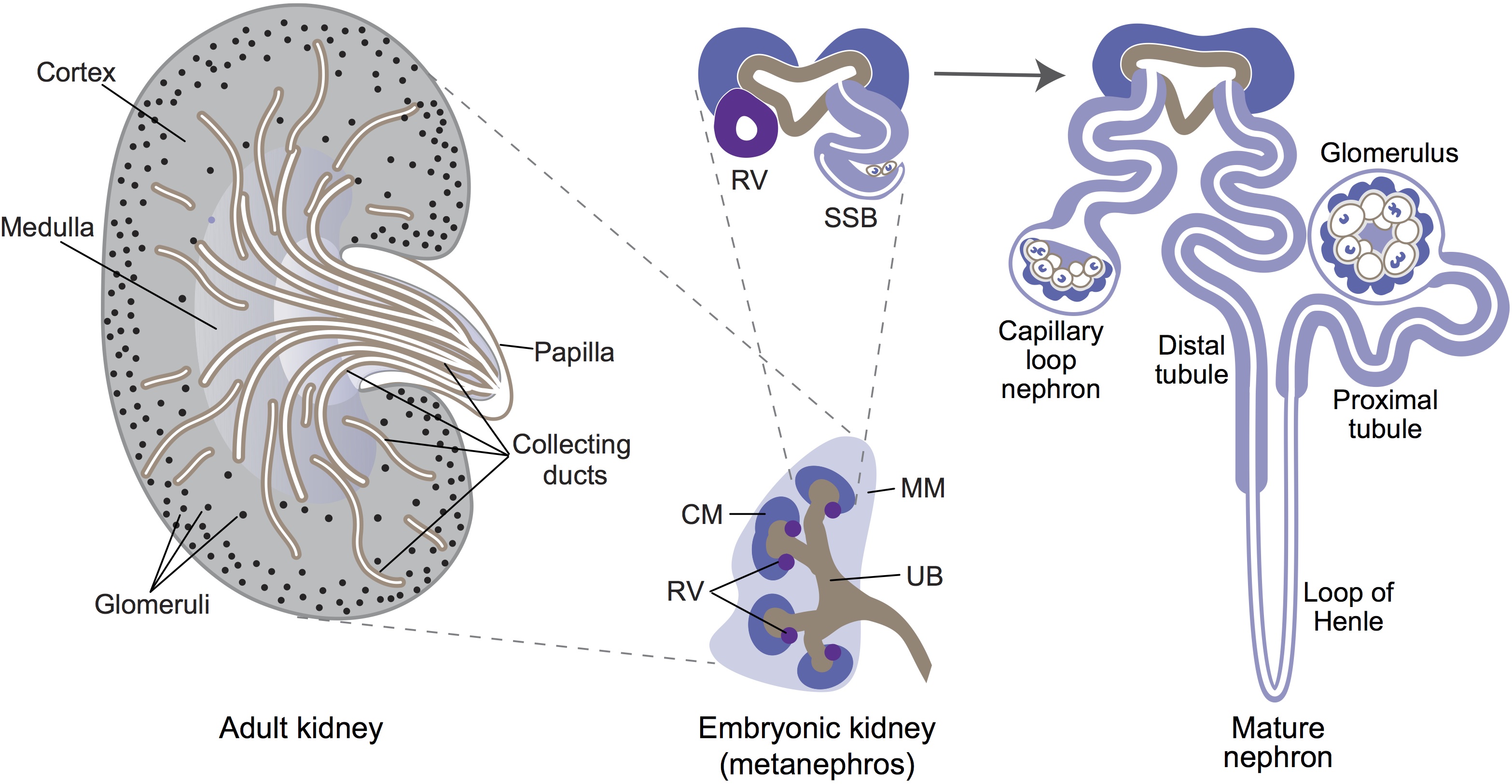 Minoru Takasato and Melissa Little discuss recent advances in the directed differentiation of pluripotent cells into kidney tissues, and how these inform our understanding of human renal development. See the Meeting Review on p.
Minoru Takasato and Melissa Little discuss recent advances in the directed differentiation of pluripotent cells into kidney tissues, and how these inform our understanding of human renal development. See the Meeting Review on p.  Elke Ober and Anne Grapin-Botton review the recent Keystone conference on Endoderm Lineages in Development and Disease, which took place last February in Colorado, USA, highlighting recent major advances in the field. See the Meeting Review on p.
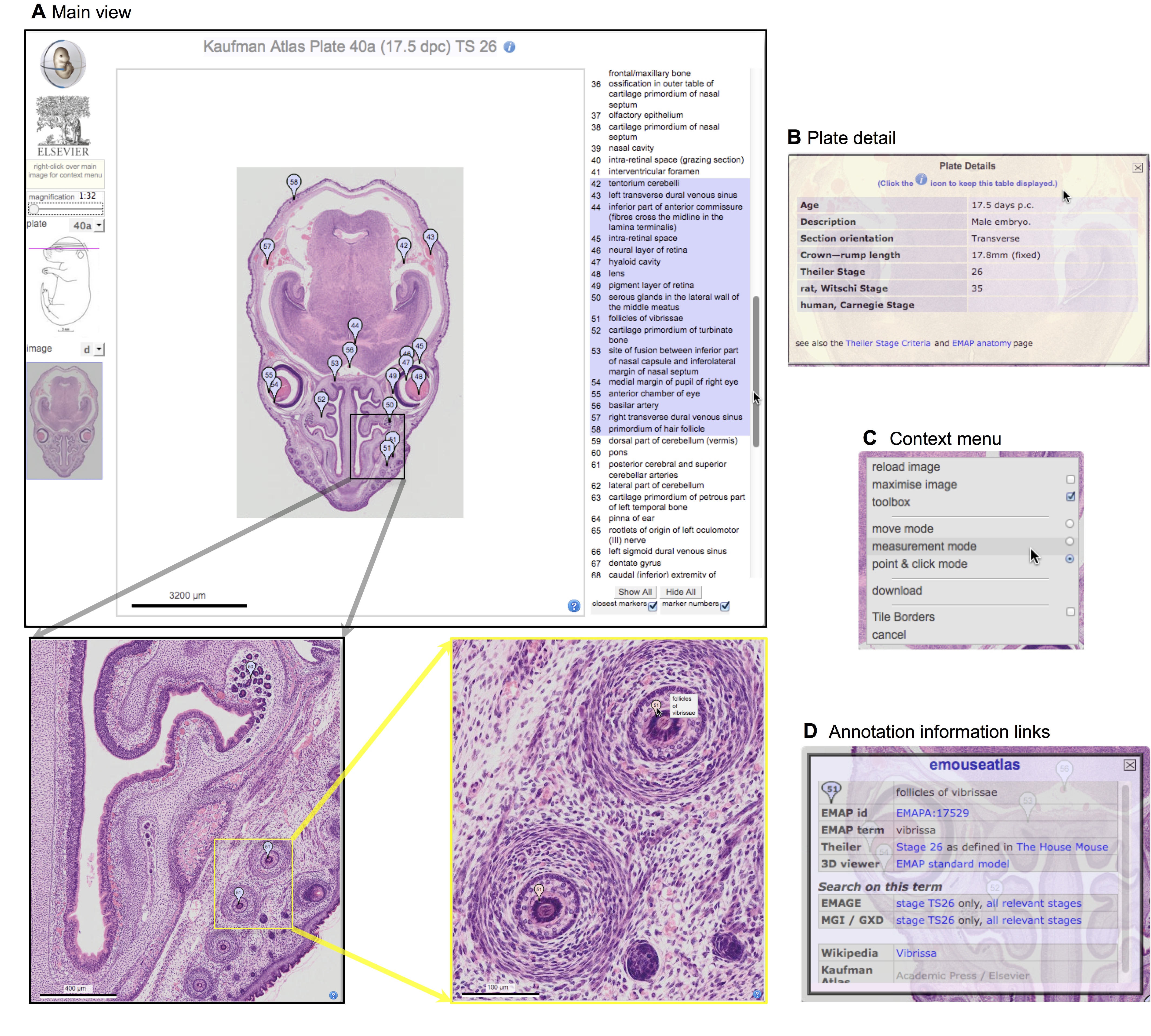
Elke Ober and Anne Grapin-Botton review the recent Keystone conference on Endoderm Lineages in Development and Disease, which took place last February in Colorado, USA, highlighting recent major advances in the field. See the Meeting Review on p.  Over two decades after the publication of ‘The Atlas of Mouse Development’ by Professor Mathew Kaufman, Baldock and colleagues announce a new online resource that provides free access to the histological images and their annotations. See the Spotlight on p.
Over two decades after the publication of ‘The Atlas of Mouse Development’ by Professor Mathew Kaufman, Baldock and colleagues announce a new online resource that provides free access to the histological images and their annotations. See the Spotlight on p.