A day in the life of a lizard lab
Posted by NoelineS, on 22 May 2015
Welcome to the Eublephosphere!
Eublephosphere (noun) – A place in which those who study the leopard gecko (Eublepharis macularius) reside.
Our names are Noeline Subramaniam and Kathy Jacyniak and we are Master’s students at the University of Guelph in Ontario, Canada. Under the supervision of Dr. Matthew Vickaryous, we have become budding regenerative biologists! Our research laboratory investigates the most fundamental question in biology: why are some tissues and organisms able to regenerate, whereas others cannot? Our model for these investigations is the leopard gecko (Eublepharis macularius), a popular reptile in the pet trade.
Research Focus
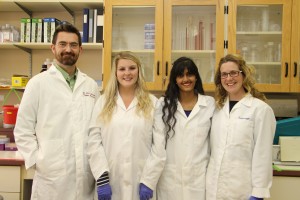
Figure 1: Members of the Vickaryous lab from left to right: Dr. Matt Vickaryous, Kathy Jacyniak, Noeline Subramaniam and Emily Gilbert
Ongoing research in our lab aims to identify and understand the biological mechanisms that permit and promote scar-free wound healing and regeneration.Emily, a PhD candidate, is examining the role of neural stem/progenitor cells in the spinal cord and the brain in response to regeneration, Noeline is investigating the role of angiogenesis in scar formation, and Kathy is exploring the role of cardiac stem/progenitor cells in the heart. If you would like to learn more about the research conducted by the Vickaryous lab, check out our website: http://www.vickaryouslab.com/. You can also follow us on Twitter @VickaryousLab, including our infamous #FluorescentFridays.
Figure 2: Dorsal view of a gecko with a beautiful original tail. This gecko was an environmental control for one of our previous studies.
Why lizards?
Many species of lizards are able to self-detach a portion of their tail to avoid predation, and then regenerate a replacement. Tail detachment, or caudal autotomy (‘self-cutting’), is a dramatic – and somewhat deceptive – phenomenon. For most species (including leopard geckos), the tail is autotomized within a vertebra at a location known as a fracture plane (essentially a non-mineralized gap in the vertebral centrum). In addition, the main arterial supply to the tail (the caudal artery) develops a regular series of muscular sphincters, with one sphincter located in advance of each fracture plane. Therefore, the site of tail loss rarely bleeds more than a couple of drops. Tail regeneration then begins spontaneously following caudal autotomy.
Why leopard geckos?
Figure 3: The leopard gecko (Eublepharis macularius) is the model we use in our lab. They can spontaneously regenerate their tail following tail loss.
Although many lizards can regenerate their tails, the husbandry requirements for some species can be challenging (e.g., the lizards are aggressive or shy, have narrow ranges of preferred temperature and humidity, particular and unusual dietary and habitat preferences, and/or are intolerant of handling). As a result, early work on lizard tail regeneration tended to focus on a few types of locally available (and wild caught) species kept over short periods of time. However, with the increasing popularity of reptiles a pets, some lizards – including leopard geckos – are now captive bred and (at least in North America) widely available. Leopard geckos are hardy, accept being handled, and have relatively simple husbandry requirements: a plastic enclosure similar in size to those used for rats and mice; a water dish; a couple of hide boxes; a heat pad under one end (to establish a thermal gradient – leopard geckos are ectotherms); and escape-proof lid (see below).
Leopard gecko fun facts:
– Caudal autotomy is not necessary for regeneration. Tails surgically amputated outside the fracture plane still regenerates
– They are members of the Eublepharidae – the ‘true eyelid’ geckos. Unlike other geckos, they have movable eyelids
– They are native to parts of Pakistan, Afghanistan, India and possibly Iran
– They have temperature-dependant sex determination – the temperature of the environment during the embryonic period helps determine the sex of the offspring
A typical day in the Vickaryous lab:
We typically start the day with a cup of coffee/tea and a discussion about our experimental plans for the day. We manage our own leopard gecko colony, and everyone has to pitch in with the daily feedings and weekly weigh and measure adventures. After the geckos have been attended to there is a good chance someone in the lab will be serially sectioning and staining tissue. Not only do we love our histology, but learning to section seems to be a rite of passage in our lab…
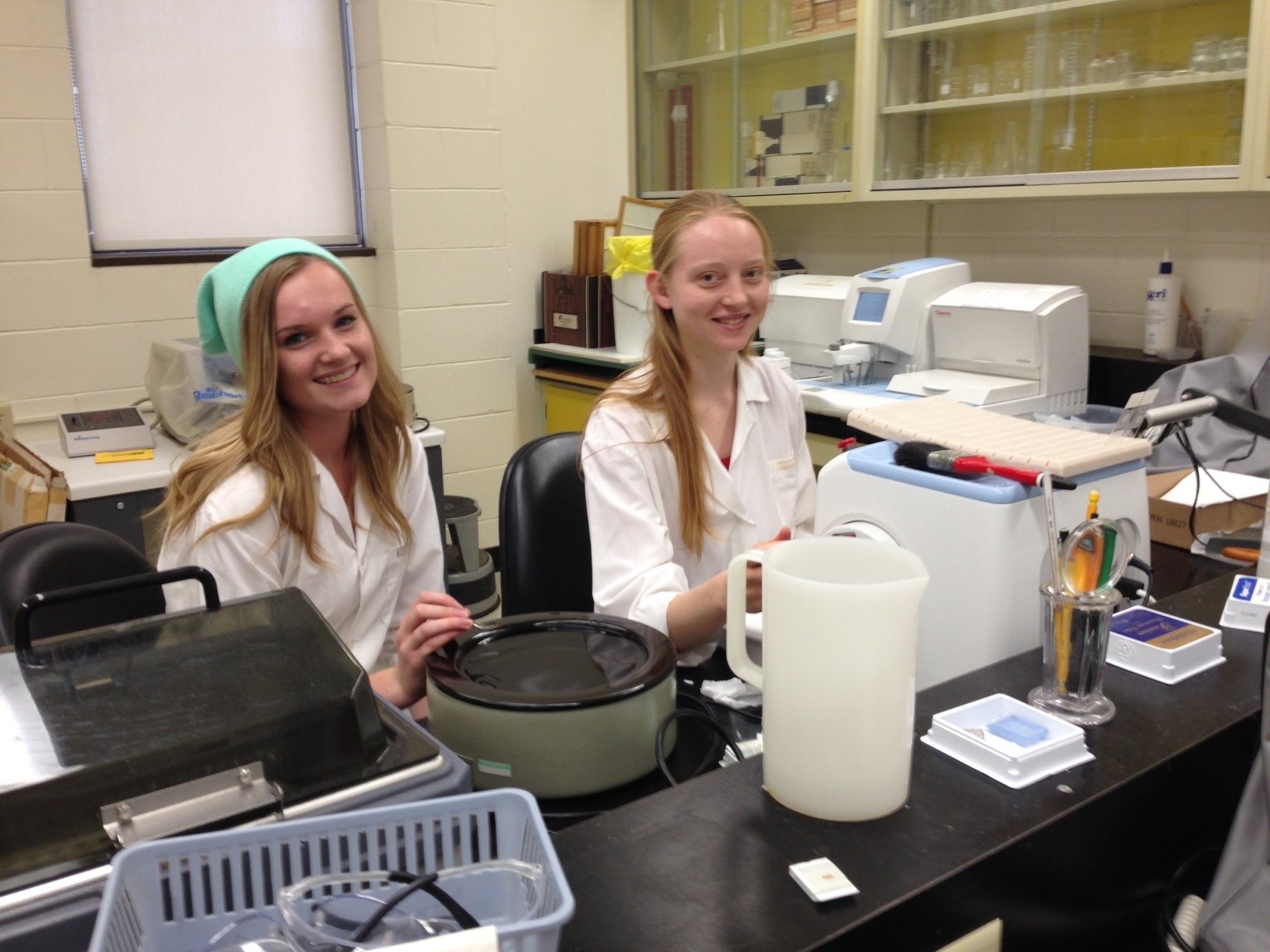
Figure 4: Undergraduate students Rebecca McDonald (left) and Alaina Macdonald (right) working in our histology prep lab. The majority of our time spent here includes processing, embedding, sectioning and staining tissues we use to study regeneration
Feeding, housing and maintaining leopard geckos:
Our gecko colony is maintained in an environmental chamber (about 28˚C) here at the University of Guelph. Leopard geckos are not terribly social, so we house each one separately – this also makes keeping track of who’s who much easier. Our gecko enclosures are roughly 36cm x 22cm x 22cm in size, and topped by a perforated lid. Although leopard geckos lack subdigital adhesive pads, they have small claws and are capable climbers (and aspiring escape artists). Fortunately, they seem to prefer showing off their talents rather than following through with the getaway.
Leopard geckos eat live prey, and we feed ours mealworms (larval Tenebrio molitor, the mealworm beetle) dusted with a calcium and vitamin D3 (cholecalciferol) powder. The use of calcium and vitamin D3 is necessary in that it helps prevent metabolic bone disease, a nasty group of disorders that can lead to brittle or distorted limb elements and jaws, paralysis and reduced growth.
Weekly duties include cleaning and changes cages, and weighing and measuring geckos. Both require a certain degree of gecko wrangling skills, and tolerance of gecko urine. Although leopard geckos are typically docile and easy to handle, they will occasionally let loose a stream – especially if you forget to wear a lab coat.
Figure 5: Aerial view of a standard gecko enclosure. Features include: two small huts and a bowl of water.
Impact of ongoing research:
Tail replacement by leopard geckos is a striking example of a naturally evolved mechanism of multi-tissue regeneration. As one of the closest living relatives to mammals, leopard geckos provide a powerful platform to study the biology of regeneration, with numerous biomedical implications. Unlike other species, leopard geckos (and other lizards) can voluntarily self-detach the tail and have a variety of structural adaptations to minimize (or at least localize) tissue damage. Thus, autotomy-mediated tail loss is arguably a less-invasive alternative to amputation to initiate the regeneration program.
Follow us on Twitter: @VickaryousLab
Website: http://www.vickaryouslab.com/
Figure 6: Tail regeneration (in dorsal view). Left to right: the original tail, two regenerating tails and variation in two complete regenerate tails.
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.






 (10 votes)
(10 votes)

 (No Ratings Yet)
(No Ratings Yet)