BSDB Gurdon Summer Studentship Report (3)
Posted by BSDB, on 3 February 2015
In 2014, the British Society of Developmental Biology (BSDB) has initiated the Gurdon Summer Studentship program with the intention to provide highly motivated students with exceptional qualities and a strong interest in Developmental Biology an opportunity to engage in practical research. The 10 successful applicants spent 8 weeks in the research laboratories of their choices, and the feedback we received was outstanding. Please, read the student reports, kindly sent to us by George Hunt.
 Modelling Developmental Neurological Disorders and Childhood-Onset Epilepsy in Caenorhabditis elegans
Modelling Developmental Neurological Disorders and Childhood-Onset Epilepsy in Caenorhabditis elegans
During the summer of 2014 I was a recipient of a Gurdon Summer Studentship awarded by the BSDB. The studentship provided the opportunity to undertake a research project in the Laboratory of Ian Hope at the University of Leeds where I study Biology.
Abnormalities in the development of the central nervous system have been implicated in the epilepsy group of chronic neurological disorders. These disorders affect fifty million people worldwide and are characterized by spontaneous recurrent seizures resulting from excessive, synchronous neuronal activity [1,2]. Improving our understanding of the biomolecular basis of epilepsy is of great importance in the search for new therapeutics. Caenorhabditis elegans provides a genetically amenable, and experimentally tractable system to model disease-relevant mutations and was the focus of this study.
Homo sapiens KCNT1 encodes a sodium-activated potassium channel that is widely expressed in the central nervous system and mutations of KCNT1 have been identified in patients with drug-resistant childhood-onset forms of epilepsy. The C. elegans KCNT1 orthologue slo-2 is widely expressed in both neurons and muscle cells [3] where it is activated during hypoxia-like physiological conditions by raised concentrations of chloride and intracellular free calcium [4]. Protein alignment identified KCNT1 amino acid residues implicated in childhood-onset epilepsy that are conserved from H. sapiens to C. elegans SLO-2. These KCNT1 variants, R474H and R928C, are associated with malignant migration partial seizures in infancy (MMPSI) [5] and autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) [6], respectively.
In order to generate R474H and R928C epilepsy-associated variants of C. elegans slo-2, equivalent mutations were introduced using the CRISPR-Cas9 system [7] to produce novel transgenic strains which can then be used to study the role of these mutations in the development of epilepsy.
Synthetic guide RNAs (sgRNA) were used to direct Cas9 nuclease activity to regions of the slo-2 locus in close proximity to the target mutational sites. Cas9-mediated cleavage generates double-stranded DNA breaks (DSBs) that are repaired in vivo by either homology-directed repair (HDR), using a homologous template, or by non-homologous end joining (NHEJ), which generates short insertion/deletion (indel) mutations. To incorporate the epilepsy-linked point mutations into the C. elegans genome, worms were microinjected with plasmid vectors expressing the modified sgRNAs and Cas9 alongside slo-2 fragments that had been PCR mutagenized to include the desired point mutations. This generated DSBs at the slo-2 locus followed by HDR to mend the DSB and introduce the desired point mutations.
 From fifteen F0 hermaphrodites injected, I obtained twenty-eight transgenic F1 individuals. Ten of these produced transgenic progeny and ten independent transgenic strains were established of which six had been targeted with the R474H equivalent alteration and four with the R928C equivalent. Microinjected DNA forms large extra-chromosomal arrays that are not transmitted to all progeny in a brood and the rate of successful CRISPR-Cas9 alterations in C. elegans varies depending on target loci [7]. To screen transgenics for alterations of slo-2, I performed single worm PCR on progeny of transgenic individuals to amplify slo-2 fragments containing the target mutation loci and assayed fragment length for the presence of indels by agarose gel electrophoresis. Following CRISPR-Cas9 generation of a DSB, activation of the NHEJ repair pathway can produce short indels at the repaired loci, providing a useful method for confirming successful CRISPR-Cas9 activity. Smaller than expected slo-2 fragments were detected in the progeny of transgenic individuals from three different strains; however, further extensive screening is required to determine whether these represent deletions within slo-2 or non-target amplification. Following confirmation of the efficacy of the CRISPR-Cas9 system the next step will be sequencing of slo-2 to identify mutants with the desired epilepsy-linked point mutations.
From fifteen F0 hermaphrodites injected, I obtained twenty-eight transgenic F1 individuals. Ten of these produced transgenic progeny and ten independent transgenic strains were established of which six had been targeted with the R474H equivalent alteration and four with the R928C equivalent. Microinjected DNA forms large extra-chromosomal arrays that are not transmitted to all progeny in a brood and the rate of successful CRISPR-Cas9 alterations in C. elegans varies depending on target loci [7]. To screen transgenics for alterations of slo-2, I performed single worm PCR on progeny of transgenic individuals to amplify slo-2 fragments containing the target mutation loci and assayed fragment length for the presence of indels by agarose gel electrophoresis. Following CRISPR-Cas9 generation of a DSB, activation of the NHEJ repair pathway can produce short indels at the repaired loci, providing a useful method for confirming successful CRISPR-Cas9 activity. Smaller than expected slo-2 fragments were detected in the progeny of transgenic individuals from three different strains; however, further extensive screening is required to determine whether these represent deletions within slo-2 or non-target amplification. Following confirmation of the efficacy of the CRISPR-Cas9 system the next step will be sequencing of slo-2 to identify mutants with the desired epilepsy-linked point mutations.
The transgenic and mutant strains will provide a useful resource in further studies of the biomolecular basis of drug-resistant childhood-onset epilepsies as they should allow production of the specific genomic modifications sought in slo-2. The effects of the slo-2 mutations on the functioning of the C. elegans neuromuscular system would then need to be characterized and findings could provide improvements in our understanding of how specific KCNT1 variants give rise to epilepsy. This research will contribute to an existing network of KCNT1 research currently being undertaken by Jonathan Lippiat and Steve Clapcote at the University of Leeds.
Overall, the BSDB Gurdon Summer Studentship provided a great opportunity to experience working in a professional research laboratory, and has strongly reinforced my desire to pursue a career in research.
References:
[1] Kwan, M.D. Schachter, S.C. and Brodie, M.J. (2012) Drug-Resistant Epilepsy. The New England Journal of Medicine 365, 919-926
[2] Engel, J.E.R. (2013) Seizures and Epilepsy (2nd Edition), New York: Oxford University Press
[3] WormBase web site (2014) Release: WS244 — [LINK]
[4] Santi, C. et al. (2003) Dissection of K+ currents in Caenorhabditis elegans muscle cells by genetics and RNA interference. PNAS 100, 14391-14396
[5] Barcia, G. et al. (2012) De novo gain of function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nature Genetics 44, 1255-1259
[6] Heron, S.E. et al. (2012) Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nature Genetics 44, 1188-1190
[7] Friedland, A,E. et al. (2013) Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature Methods 10, 741-743


 (6 votes)
(6 votes) – Christoph and his colleagues wrote about their recent Development paper, where they
– Christoph and his colleagues wrote about their recent Development paper, where they 

 (No Ratings Yet)
(No Ratings Yet)





 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series 
 (1 votes)
(1 votes)

 Invasive cells such as immune and metastatic cancer cells form protrusions known as invadosomes, which mediate adhesion to the underlying substrate and induce extracellular matrix degradation – thus promoting invasiveness. On p.
Invasive cells such as immune and metastatic cancer cells form protrusions known as invadosomes, which mediate adhesion to the underlying substrate and induce extracellular matrix degradation – thus promoting invasiveness. On p.  For many years it has been clear that embryonic cells from different mouse strains differ in their properties for generating embryonic stem cells (ESCs). Specifically, ESCs can be generated and maintained from some (‘permissive’) strains in the presence of serum and LIF, whereas these conditions are insufficient to support self-renewal of cells derived from other genetic backgrounds (‘non-permissive’ strains). Here (p.
For many years it has been clear that embryonic cells from different mouse strains differ in their properties for generating embryonic stem cells (ESCs). Specifically, ESCs can be generated and maintained from some (‘permissive’) strains in the presence of serum and LIF, whereas these conditions are insufficient to support self-renewal of cells derived from other genetic backgrounds (‘non-permissive’ strains). Here (p. 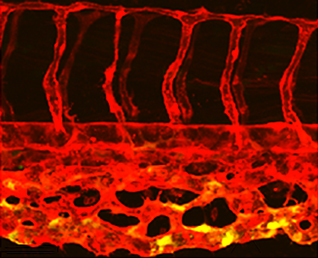 β-catenin is a multifunctional protein that acts both as a downstream mediator of the Wnt signalling pathway and as a core component of adherens junctions. It is also widely expressed during development. Consequently, dissecting out the specific functions of β-catenin in particular contexts can be challenging. Shigetomo Fukuhara, Naoki Mochizuki and colleagues now report a transgenic zebrafish line that allows the visualisation of β-catenin activity in living tissues (p.
β-catenin is a multifunctional protein that acts both as a downstream mediator of the Wnt signalling pathway and as a core component of adherens junctions. It is also widely expressed during development. Consequently, dissecting out the specific functions of β-catenin in particular contexts can be challenging. Shigetomo Fukuhara, Naoki Mochizuki and colleagues now report a transgenic zebrafish line that allows the visualisation of β-catenin activity in living tissues (p. 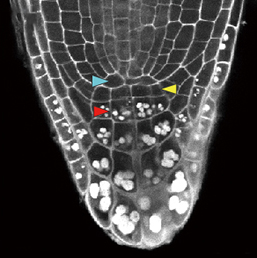 In the plant root meristem, a highly orchestrated pattern of cell divisions controls both root growth and cell fate. A large number of signalling factors and transcriptional regulators have been found to control proliferation and differentiation in the root meristem, including small peptide ligands of the CLV3/CLE family. However, the mechanisms by which these peptides act remain poorly understood. Now (see p.
In the plant root meristem, a highly orchestrated pattern of cell divisions controls both root growth and cell fate. A large number of signalling factors and transcriptional regulators have been found to control proliferation and differentiation in the root meristem, including small peptide ligands of the CLV3/CLE family. However, the mechanisms by which these peptides act remain poorly understood. Now (see p. 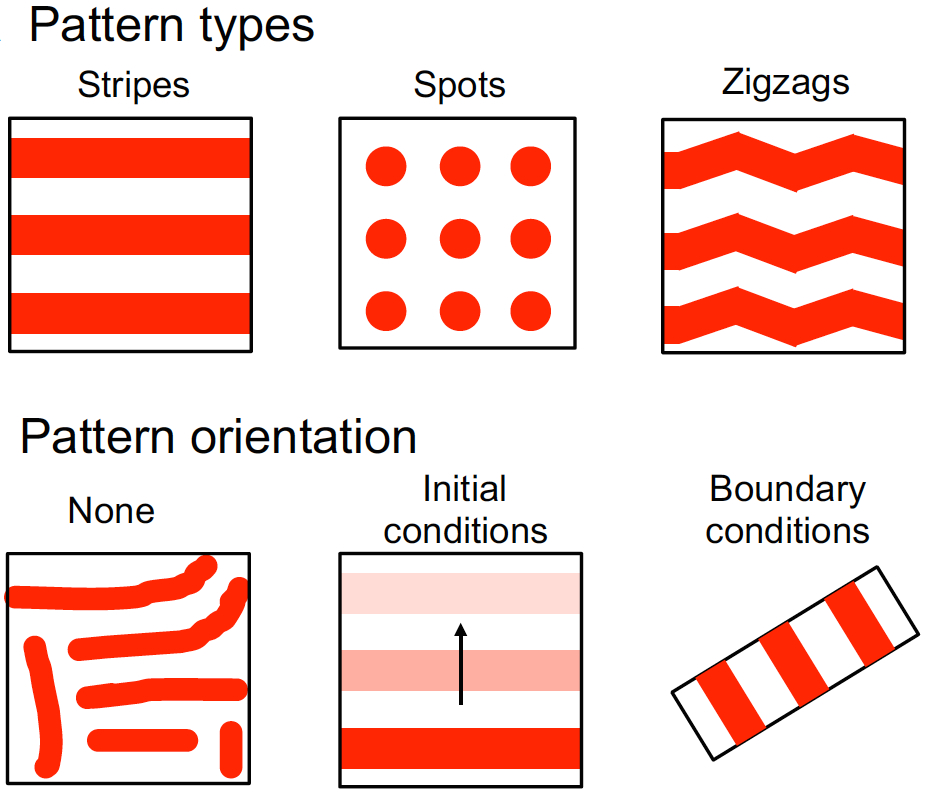 Hiscock and Megason present a mathematical approach to understanding periodic patterning development. They also suggest ways in which different types of model can be tested, illustrating the potential of this methodology using specific biological examples. See the Hypothesis on p.
Hiscock and Megason present a mathematical approach to understanding periodic patterning development. They also suggest ways in which different types of model can be tested, illustrating the potential of this methodology using specific biological examples. See the Hypothesis on p. 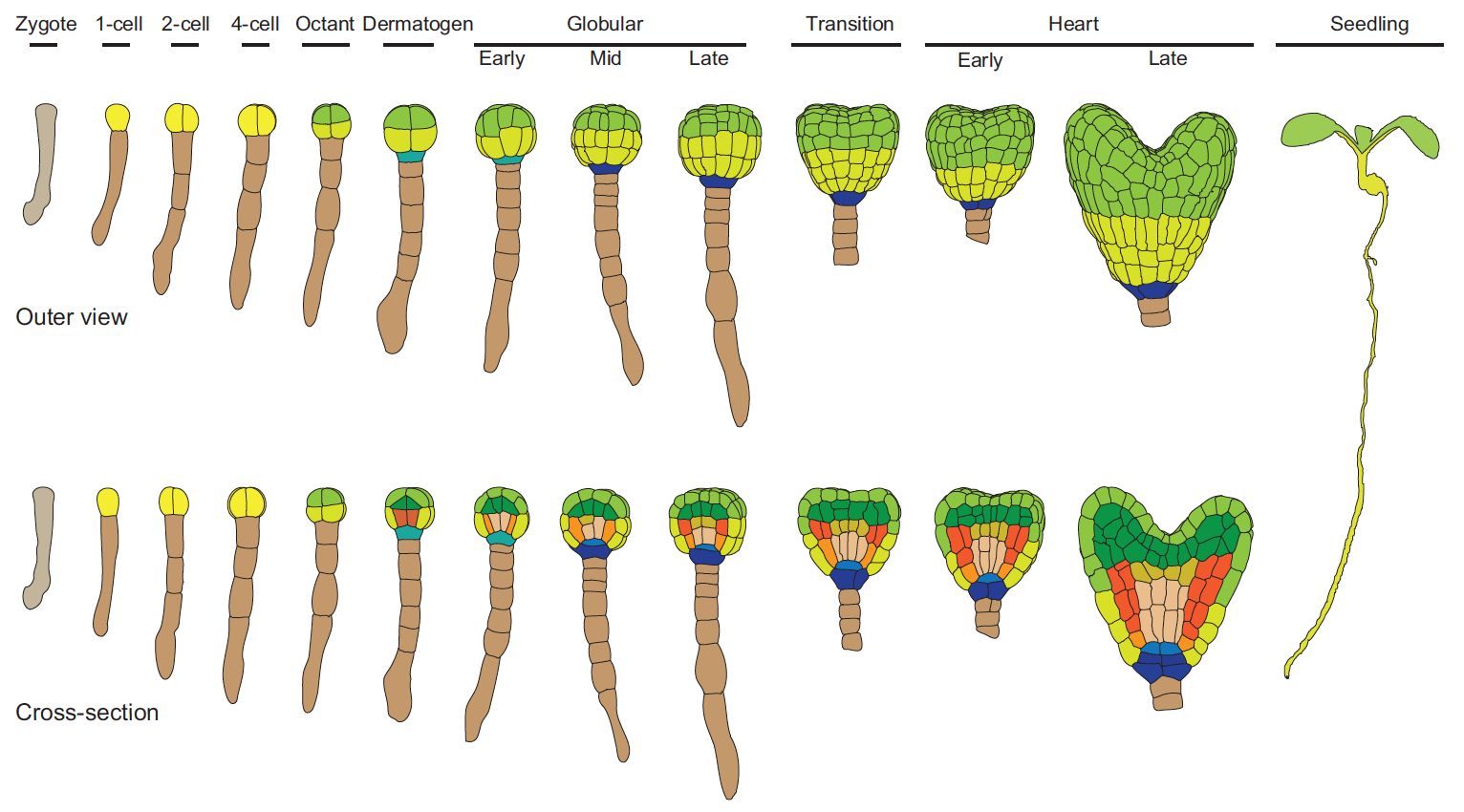 The cell fate decisions and patterning steps that occur during plant embryogenesis are essential reiterated during organogenesis. Hove, Lu and Weijers summarise our current understanding of the early stages of plant embryogenesis, with a focus on how the major lineages are specified. See the Review on p.
The cell fate decisions and patterning steps that occur during plant embryogenesis are essential reiterated during organogenesis. Hove, Lu and Weijers summarise our current understanding of the early stages of plant embryogenesis, with a focus on how the major lineages are specified. See the Review on p.