A typical day spent in our lab’s aquarium room will find me soaked, top-to-bottom, in seawater. The other members of my lab seem not to have this issue, but I’ve always seemed to get “in” to my science, literally, when it comes to these types of chores. The trouble with marine models is that they require salt water, and lots of it. This requires that I balance half of my small frame over the edge of a 44 gallon drum so that I may reach to stir in any last bits of un-dissolved salt. I pipette a small drop of the freshly made salt water onto the spectrometer, closing one eye as I bring the device to the other, pointing it up to the light in order to check that the salinity is within 28 – 32 PPT. This always makes me feel like a pirate. Normally, the day-to-day care of the stock tanks is hurried time away from research, but on the days of failed experiments, these mundane tasks—making seawater, cleaning tanks, fixing pumps, and the like—can be a satisfying reprieve. Until, that is, you get a giant mouthful of dirty seawater while attempting to start the flow of an aquarium siphon to vacuum the bottom of the tanks.
When I’m not maintaining the tanks in the pleasantly warm and humid aquarium room, I can be found in the back corner of the lab, carefully manipulating delicate embryos under the microscope with homemade tools; at my bench, changing wash after wash of an immunofluorescent antibody stain; or in the tiny windowless room filled predominantly by the very expensive confocal microscope, watching the attached computer screen hopefully as bright layers of color appear, digital slice by digital slice. The images that form are the visualized results of weeks of work. Until you actually see your results, anything is possible, claimed one past mentor; I’m not sure how much I buy into this “Schrödinger’s results” mindset, but I’m sometimes afraid to look just the same.
A new model
Par … hi … allie? The name of this up-and-coming model organism doesn’t quite roll off the tongue, and I have yet to meet someone who can pronounce Parhyale for the first time without stumbling. Parhyale hawaiensis, perhaps better known as “beach hoppers” or “sand fleas”, are amphipod crustaceans that like to hang out on decaying plant matter in intertidal, equatorial waters worldwide, or, in our lab, on carrots and air bubblers.
If you haven’t had the chance to meet (or mispronounce) this up-and-coming model organism, don’t worry—you will, because these guys are making quite the splash in the field of developmental biology. When I first joined Parhyale pioneer Nipam Patel’s lab as an incoming graduate student at UC Berkeley, I knew nothing of them. I came from a vertebrate-centric background, and had little experience with arthropods (or anything not warm and fuzzy, for that matter). So green was I to the frequently cited arthropod orders, that when I first heard the term “amphipod” I thought I was mishearing the term “arthropod”. Parhyale were my first introduction to this new and wonderful world of invertebrates, and I would soon learn just how important their appendages are for answering questions about morphological diversity. Indeed, the phylum Arthropoda means “jointed foot” and amphipod refers to the two different orientations of the thoracic appendages in this order.
Using Parhyale, our lab studies developmental mechanisms that pattern the body plan and the evolutionary changes that have led to the diversity of arthropod appendages. With their amiability to laboratory manipulation, total cleavage forming stereotyped lineages early in development, and boundless supply of embryos, Parhyale make an excellent model for developmental studies.
Tupperwares of love
The first thing you may notice as you enter our lab is the hum of the air bubblers from the Parhyale tanks—clusters of various sized Tupperware that populate each bench. The tanks soon fall into the background drone of freezers and other electronic equipment. The tanks vary in cleanliness (this may or may not be correlated to the availability of undergrads), though the Parhyale don’t seem to mind. They are detritivores, and live off of decaying organic matter in the ocean. In the lab, we feed them carrots. Yes, carrots. They cling to them in droves; presumably eating whatever it is that grows on them.
Indeed, that’s what Parhyale were chosen for—their hardiness. When Bill Browne (then a graduate student in the Patel lab) went to Chicago’s Shedd Aquarium in search of new crustaceans for the lab, he didn’t go to the display tanks. Rather, he surveyed the filtration system and discovered an ideal lab animal—one that survives on garbage without the need for constant care. This hardiness can be attributed to the constant, naturally occurring temperature and salinity fluctuations of the shallow water and intertidal habitats they have evolved to flourish.
But Parhyale offers more than just hardiness and ease of bench top maintenance. They offer fecundity; it’s part of their charm. “Aw, look at all of the mommies swimming around paired with their babies!” exclaimed one colleague when I first showed her my research subjects. No, I had to explain, when a boy Parhyale loves a girl Parhyale, the larger male shows his affections by dragging her around with him in pre-mating “amplexus” until she molts and he fertilizes the eggs and releases her. Thus begins a new cycle of embryo production. Development labs are necessarily centered to the reproductive agenda of their animals, but there is no finickiness—or shortages of gravid females—in our Tupperwares of love. Females can produce embryos every two weeks after (quickly) reaching sexual maturity, and, as tropical species tend to do, they produce broods year round. To learn more about these guys, check out The crustacean Parhyale hawaiensis: A New Model for Arthropod Development.
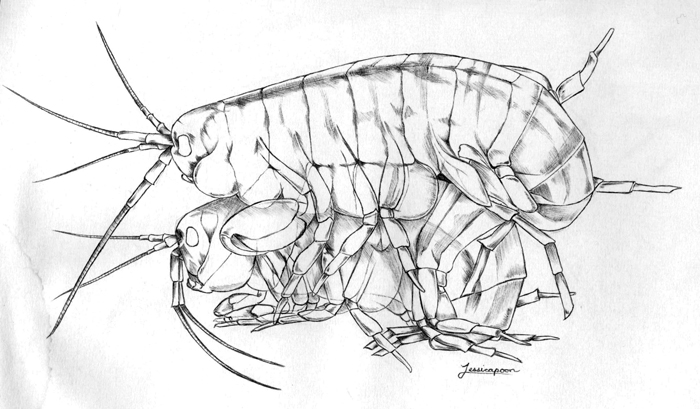
Parhyale Amplexus. Drawing by Jessica Poon.
A day in the life of a Parhyale wrangler
On any given day, my tanks contain a good mix of males, females, mate pairs, and gravid females. But I am after something specific—one- and two-cell embryos. To increase my chances of catching this four to nine hour window of a 10-day embryogenesis, I go for the pairs. I cut the end off of a 3 mL transfer pipette so that it’s wide enough (barely) to suck up a mating pair, and, as I eye my quarry among the carrots, pH-buffering gravel, and non-paired individuals, let the rodeo begin. I’m good at this now; people new to the lab are impressed with the quickness at which I can pluck my chosen paired Parhyale out of the water, despite their fast reactions and expert swimming. These pairs are placed in a separate tank and regularly observed for detachment. Newly single males are removed, and the freshly gravid females are detained for embryo collection; adding a few microliters of clove oil to their water will knock them out.
Parhyale, like all amphipods, carry their embryos in a brood pouch on their ventral side. Delicately clasping the sleeping body with forceps, I use the smooth end of a glass pipette pulled and rounded over a flame to gently scrape along her ventral pouch; the embryos pop up and drift away. Done correctly, embryo collection is fairly non-invasive for the female. The clove oil anesthetic soon wears off and the females can be returned to their tanks to resume their breeding cycle.
The one and two-cell embryos are prime for microinjection. siRNA against my gene of choice is loaded into a microinjection needle pulled from a small capillary tube. The loud “WHAP” made by the force of the needle puller separating as a pulled capillary tube breaks in two still makes me jump. My dedicated undergraduate, Jennifer Wang, does the lion’s share of the siRNA injections that will knockdown the targeted mRNA. If we want to knock down expression throughout the entire animal, we inject the one-cell embryo, or both cells at the two-cell stage. For some experiments, we can also achieve lineage specific knockdown by injecting one or more cells at the eight-cell stage.
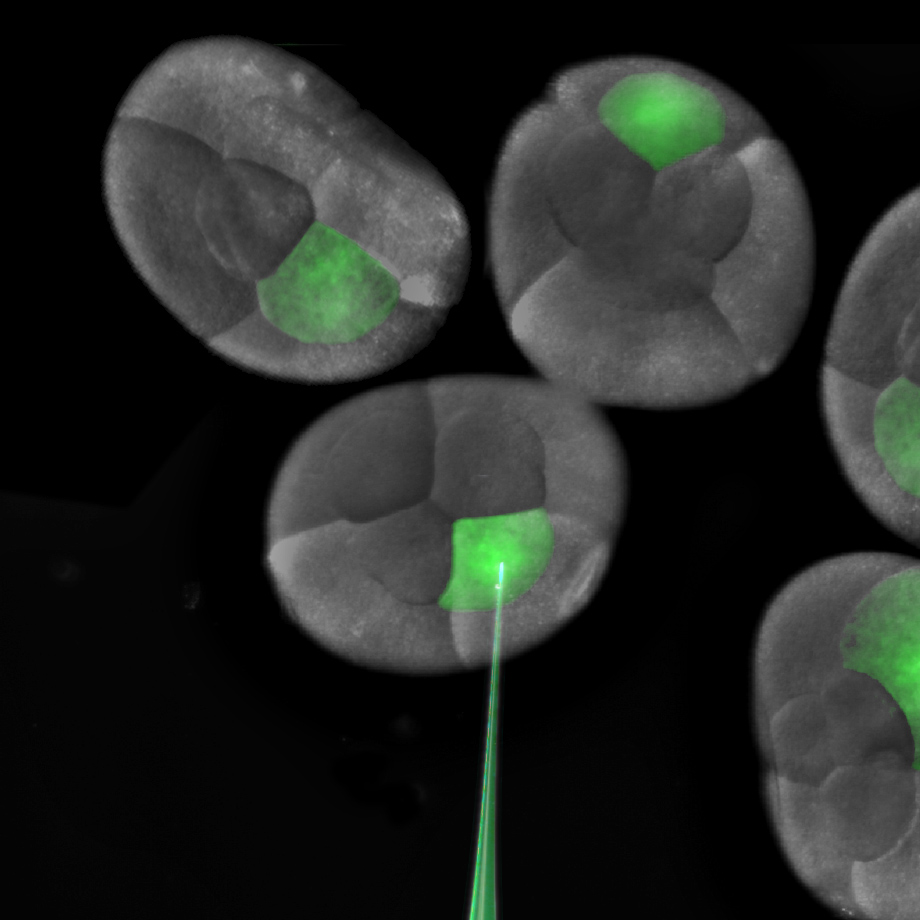
Injection of a single blastomere of an eight cell embryo.
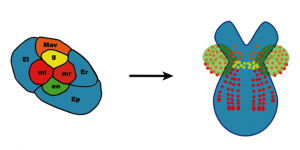
Fate map of the eight cell stage of Parhyale development.
Once injected, this is when we have to really care for them. Parhyale embryos will develop just fine in a tissue culture dish, but the water must be changed several times a week to avoid fungal contamination and death. It’s easy to keep track of embryonic stages in Parhyale thanks to the detailed Parhyale staging system of Browne, et al. There are 30 stages divided by time point and visual characteristics, many of which have lab nicknames names such as “frosted-soccer ball” stage or simply “leggy”, which is the stage I’m after.
Now comes the fun part, the part where I sit for hours dissecting out embryo after embryo from their extra-embryonic membranes. Learning to dissect out an embryo—intact—requires countless hours of frustration at the beginning, but it’s rewarding, and, eventually (dare I say)—fun, once you get the hang of it. To accomplish these tiny dissections, we thread small pieces of tungsten wire into insulin needles, and sharpen these dissection needles by dipping their ends into a beaker of sodium hydroxide with a current running through it. I use these tools to secure a rolling embryo, poking a hole through both membranes to allow yolk to flow out, and the fixation solution (4% paraformaldehyde in sea water) to flow in. Teasing the embryo, the outer membrane seems to pop right off. Removing the inner membrane takes much more careful manipulation, and much more patience.
Once I have a tube full of fixed embryos with their membranes removed, I use immunofluorescent antibody stains to visualize gene expression patterns and structures within the embryos. Easy as making cookies—except that my antibody stains usually turn out more successfully than my half-hearted attempts at baking (turns out that antibody incubation times are much more flexible than bake times). The tricky part is not accidently sucking up a small (sometimes floating) embryo while performing wash after wash in PBS with Triton. The last step is clearing with glycerol; the embryos are now ready to be mounted. I mount one Parhyale embryo per slide, carefully splaying out each limb and using the twisted edge of a Kimwipe to suck up any excess glycerol. I hold my breath as I place the cover slip over a particularly nice mount—it’s always the perfect ones that seem to get messed up during this step. If I am lucky, I can snag an opening on the confocal; we share the microscope with the entire floor, and scheduling can be tight. I’m in luck, there’s an evening slot. With no one behind me, I’m able to confocal late into the night. As the confocal scans, the gene expression patterns are slowly revealed, ventral to dorsal, in a satisfying mix of my chosen colors: aqua, neon green, Christmas red, and magenta bordering on pink.
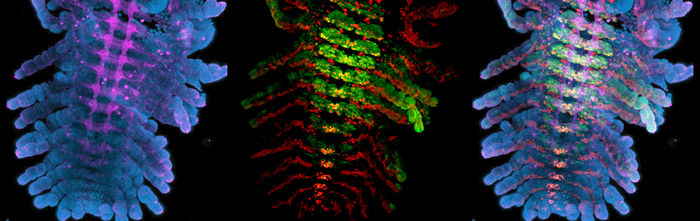
Immunofluorescent stains of Parhyale embryos.
Left: Blue = DAPI, Magenta = anti-HRP (nervous system). Center: Red = Engrailed, Green = Ultrabithorax. Right: Images combined
Right now I am focused on the function of homeotic genes in crustaceans, and how evolutionary changes in these genes have contributed to the diversification of arthropod appendages. I use siRNAs to knock down individual Hox genes, and then use various markers to examine changes in the ectoderm, nervous system, and musculature to see if all parts of the segment are transformed to understand how the fate of these different structures is coordinated during development. As I make my way through my 3rd year as a graduate student, I now consider myself one of the few, the proud, the Parhyale researchers. My friends (the non-sciency ones) consider me unemployed.
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
 (16 votes)
(16 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)




 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series  (16 votes)
(16 votes)
 (9 votes)
(9 votes) Hox genes are classically known for their roles in patterning the anterior-posterior axis of animals. Now, on
Hox genes are classically known for their roles in patterning the anterior-posterior axis of animals. Now, on 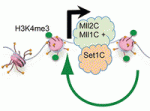 Histone H3 lysine 4 trimethylation (H3K4me3) is a universal epigenetic mark that is deposited by histone methyltransferases. This mark can be found in the context of bivalent promoters, which harbour both repressive H3K4me3 and active H3K27me3 marks and hence are thought to be poised for lineage-specific activation or repression. Here, Francis Stewart, Henk Stunnenberg and co-workers challenge this model of poising (
Histone H3 lysine 4 trimethylation (H3K4me3) is a universal epigenetic mark that is deposited by histone methyltransferases. This mark can be found in the context of bivalent promoters, which harbour both repressive H3K4me3 and active H3K27me3 marks and hence are thought to be poised for lineage-specific activation or repression. Here, Francis Stewart, Henk Stunnenberg and co-workers challenge this model of poising ( The correct architecture of dendritic trees is essential for the wiring and function of neuronal circuits. A number of cell extrinsic factors are known to regulate dendrite shape and patterning, but here Don van Meyel and co-workers show that the transcription factor Longitudinals Lacking (Lola) regulates expression of the actin nucleation protein Spire (Spir) to sculpt dendrite architecture in Drosophila (
The correct architecture of dendritic trees is essential for the wiring and function of neuronal circuits. A number of cell extrinsic factors are known to regulate dendrite shape and patterning, but here Don van Meyel and co-workers show that the transcription factor Longitudinals Lacking (Lola) regulates expression of the actin nucleation protein Spire (Spir) to sculpt dendrite architecture in Drosophila ( Morphogen and transcription factor gradients are known to pattern tissues during development, but how these gradients are established is unclear. Using mouse embryos, embryonic stem cells (ESCs) and mathematical modelling, Heiko Lickert and colleagues show that the microRNA miR-335 fine-tunes a transcription factor gradient in the endoderm (
Morphogen and transcription factor gradients are known to pattern tissues during development, but how these gradients are established is unclear. Using mouse embryos, embryonic stem cells (ESCs) and mathematical modelling, Heiko Lickert and colleagues show that the microRNA miR-335 fine-tunes a transcription factor gradient in the endoderm ( It is known that nutrient availability affects cell proliferation, but how nutrients affect the proliferation-differentiation programme of cells is unclear. On
It is known that nutrient availability affects cell proliferation, but how nutrients affect the proliferation-differentiation programme of cells is unclear. On  Variation in jaw size has been crucial to the evolution and adaptation of vertebrates. On
Variation in jaw size has been crucial to the evolution and adaptation of vertebrates. On 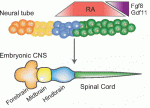 All muscle movements, including breathing, walking, and fine motor skills rely on the function of spinal motor neurons. Here, Kevin Eggan and colleagues discuss how the logic of spinal motor neuron development has been applied to generate motor neurons either from pluripotent stem cells by directed differentiation and transcriptional programming, or from somatic cells by direct lineage conversion. See the Primer on p.
All muscle movements, including breathing, walking, and fine motor skills rely on the function of spinal motor neurons. Here, Kevin Eggan and colleagues discuss how the logic of spinal motor neuron development has been applied to generate motor neurons either from pluripotent stem cells by directed differentiation and transcriptional programming, or from somatic cells by direct lineage conversion. See the Primer on p. 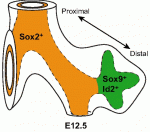 The respiratory system, which consists of the lungs, trachea and associated vasculature, is essential for terrestrial life. In recent years, extensive progress has been made in defining the temporal progression of lung development. This has led to exciting discoveries including the derivation of lung epithelium from stem cells and the discovery of new targets for therapeutics. Michael Herriges and Ed Morrisey highlight review these recent advances in our understanding of lung development and regeneration. See the Review on p.
The respiratory system, which consists of the lungs, trachea and associated vasculature, is essential for terrestrial life. In recent years, extensive progress has been made in defining the temporal progression of lung development. This has led to exciting discoveries including the derivation of lung epithelium from stem cells and the discovery of new targets for therapeutics. Michael Herriges and Ed Morrisey highlight review these recent advances in our understanding of lung development and regeneration. See the Review on p. 