Photo-Morpholinos
Posted by Philip Washbourne, on 3 April 2012
In the study of the roles of genes during development, one problem that is often faced by researchers examining ‘late’ roles of genes is one of obscuration by temporal pleiotropy. That is: gene mutations and gene knock-down obscure late roles in development, because early phenotypes, such as cell death or malformed embryos, make it impossible to examine later functions. A common way to achieve gene knock-down in developing zebrafish and Xenopus tadpoles, is by using Morpholinos (MOs). These are antisense oligonucleotides composed of morpholine-modified nucleotide subunits, commonly used to block the initiation of translation or to block splicing. We were faced with just such a pleiotropy problem with a set of genes we are currently studying. We considered trying to cage MOs ourselves, rendering them temporally activatable, by linking two complementary MOs together using a photo-cleavable crosslinker, much as has already been described previously (e.g. Shestopalov et al. 2007). We called Gene Tools LLC, the company that synthesizes MOs, to ask them whether they had any suggestions with the chemistry. Paul Morcos, at Gene Tools LLC, mentioned that they had been developing such a product for a while and thought they were close to having something worth trying out.
A few months later, Paul called, asking if we would like to test their new photo-controllable MOs. They had designed a photo-cleavable subunit with the size and chemical properties so as to ‘blend in’ with the other MO subunits, but that could be cleaved by UV light. This made it possible to design (1) sense-photo-MOs (S-photo-MOs), which would base pair with ‘regular’ MOs and thus block their activity until UV exposure, and (2) antisense-photo-MOs (AS-photo-MOs), which would function just like ‘regular’ MOs until inactivation by UV exposure. These could then be used to turn gene function off at a given time point (using S-photo-MOs mixed with complimentary MOs), or, in a novel application, turn gene function on at a given time point (using AS-photo-MOs). These MOs would then allow researchers to study early or late gene function respectively, creating a more complete picture of gene function during development; and to find the critical time period in which gene function is necessary for the proper development of specific cell populations. Furthermore, we considered that by using a laser, we could generate cell-autonomous knock-down of genes in specific cell populations, eliminating the need for cell transplant experiments from WT into mutant backgrounds, or vice versa.
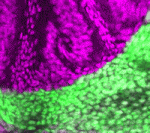
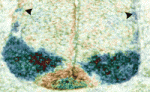
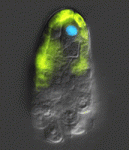
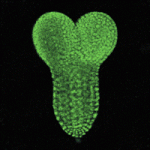
We chose ntla, sox10 and gal4 genes as targets with which to beta-test the photo-MOs. Both ntla and sox10 knock-down have easily identifiable phenotypes. Gal4 was chosen as it would potentially give researchers the additional temporal and spatial control over transgene expression for a wide variety of applications. The results are published in the latest edition of Development, showing that these new reagents from Gene Tools work surprisingly well. As with regular MOs, the ideal working concentration needs to be determined, and one needs to find the best ratio of S-photo-MO to regular MO (1:1 is a good starting point). Once optimized these reagents perform well, almost completely blocking MO activity before UV and completely releasing MO activity after UV. Interestingly, our results confirmed that ntla is only necessary as late as tailbud stage to drive medial precursor cells towards a notochord fate, and that sox10 is not required for neural crest formation, but for maintenance. Experiments with Gal4 photo-MOs allowed us to restrict expression of a transgene (in this case GFP) to primary and not secondary motoneurons. This was not possible until now using conventional transgenic approaches. We hope these tools go on to help researchers circumnavigate problems of temporal pleiotropy during development and help enhance the expression patterns of transgenes in the future.
![]() Alexandra Tallafuss, Dan Gibson, Paul Morcos, Yongfu Li, Steve Seredick, Judith Eisen, & Philip Washbourne (2012). Turning gene function ON and OFF using sense and antisense photo-morpholinos in zebrafish Development, 139 (9), 1691-1699 : 10.1242/dev.075390
Alexandra Tallafuss, Dan Gibson, Paul Morcos, Yongfu Li, Steve Seredick, Judith Eisen, & Philip Washbourne (2012). Turning gene function ON and OFF using sense and antisense photo-morpholinos in zebrafish Development, 139 (9), 1691-1699 : 10.1242/dev.075390


 (8 votes)
(8 votes) Paul O’Neill shared some
Paul O’Neill shared some 



 (No Ratings Yet)
(No Ratings Yet)