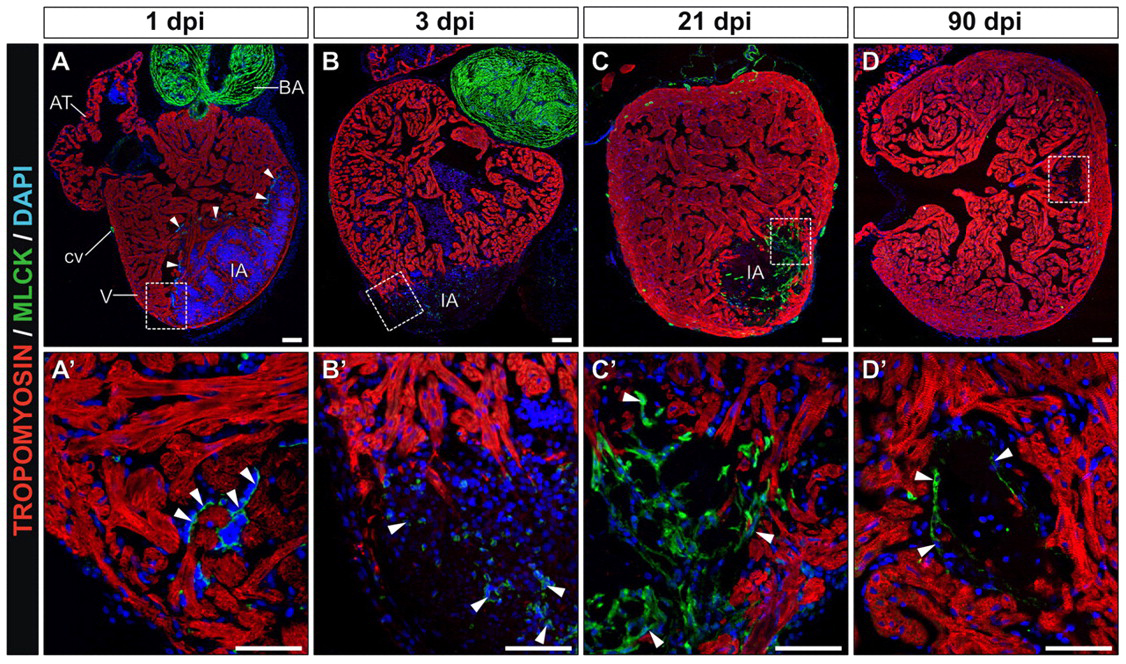
Zinc Finger Nucleases targeting genes in a frog near you!
Posted by Anita Abu-Daya, on 9 May 2011
Loss-of-function studies in Xenopus have been, until recently, limited to transient knockdowns by injection of morpholino antisense oligonucleotides. In part because of X. laevis’ complex allotetraploid genome, the system lacked techniques for targeted gene disruption. In recent years the use of the closely related Xenopus tropicalis, a true diploid with one of the smallest tetrapod genomes, has allowed the addition of genetics to the suite of molecular and embryonic techniques enjoyed by frog researchers, and in a recent PNAS paper from Richard Harland’s lab, John Young and colleagues provide the first example of targeted gene knockout in amphibians using zinc finger nucleases.

The C2H2 zinc finger, the most common DNA binding domain, was first discovered, appropriately enough in Xenopus. Each zinc finger recognizes a specific trinucleotide sequence, and several fingers can be linked in tandem to bind a longer DNA sequence, with a high degree of specificity. In addition it has been possible to engineer zinc finger protein domains with novel specificities and to couple them to the DNA cleavage domain of the restriction enzyme Fok1, giving the possibility of engineering zinc finger nuclease (ZFN) that targets a unique genomic locus and produces a double strand break (DSB). In the absence of a homologous template, cells repair DSBs by non-homologous end joining which sometimes introduces small insertions or deletions. If these result in a frame-shift, and the ZFN is targeted to the coding sequence of a gene, a null or hypomorph allele may be produced. So far ZFNs have been used to target genes in drosophila, zebrafish and rat. The Young et. al. paper has now extended the use of the technique to Xenopus.
The authors first tested whether somatic ZFN genome editing was possible in amphibian embryos by injecting mRNA encoding a nuclease targeted against GFP into eggs heterozygous for a single-copy ubiquitous GFP transgene. Lower doses of mRNA resulted in a mosaic loss of fluorescence and when the mRNA concentration was increased most cells in the resulting embryos were not fluorescent. Sequencing of the GFP locus in injected embryos showed insertions and deletions of 5-20 bp in the targeted sequence.
A panel of ZFNs targeting noggin was then designed and 6 constructs that were active in a yeast-based single stranded annealing assay were tested in Xenopus embryos. All ZFNs were tolerated well by the embryos and produced mutant amplicons at a frequency of 10-47% with insertions/deletions ranging from 5-195 bp. To test heritability, injected embryos carrying mutated loci were raised to sexual maturity and crossed to wild type frogs. 3 mutated noggin alleles were recovered in the next generation, including a 4bp insertion resulting in a premature stop codon, probably a null allele, and a 12 bp deletion which might be a hypomorph, showing that ZFNs can be used to produce an allelic series of any gene of interest.
An exciting future possible application of ZFNs is the ability of DSBs to increase the rates of homologous recombination, in the presence of template DNA with homologous ends, by many orders of magnitude. Although this has not been tested in embryos yet, it appears to work well in cell-lines, raising the possibility of using ZFNs to generate knockins as well as knockouts in genes of interest. For the time being, however, the ability to disrupt specific genes in Xenopus is an exciting development for loss-of-function studies in this model system.
![]() Young, J., Cherone, J., Doyon, Y., Ankoudinova, I., Faraji, F., Lee, A., Ngo, C., Guschin, D., Paschon, D., Miller, J., Zhang, L., Rebar, E., Gregory, P., Urnov, F., Harland, R., & Zeitler, B. (2011). Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases Proceedings of the National Academy of Sciences, 108 (17), 7052-7057 DOI: 10.1073/pnas.1102030108
Young, J., Cherone, J., Doyon, Y., Ankoudinova, I., Faraji, F., Lee, A., Ngo, C., Guschin, D., Paschon, D., Miller, J., Zhang, L., Rebar, E., Gregory, P., Urnov, F., Harland, R., & Zeitler, B. (2011). Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases Proceedings of the National Academy of Sciences, 108 (17), 7052-7057 DOI: 10.1073/pnas.1102030108


 (8 votes)
(8 votes)
 (6 votes)
(6 votes)