BSDB Gurdon Studentship Report – Xueqing Li
Posted by Xueqing Li, on 6 January 2023
The role of canonical Wnt signalling in embryogenesis of the invertebrate chordate Ciona intestinalis.
Ascidians, as the closest invertebrate sister group of vertebrates, are important to study the development and evolution of our own species. Their gene networks are closely related to those of humans, but without the complexities that were introduced via the whole genome duplications that occurred at the origin of vertebrates. A particular conserved mechanism, canonical Wnt (cWnt) signalling is essential to various development processes, especially in patterning along the anterior-posterior axis. Understanding the role of the cWnt pathway in ascidians would be helpful to elucidate the emergence of chordates and their evolution.
The summer project was complementary to ongoing work in the lab of David Ferrier (in the Scottish Ocean Institute, University of St. Andrews), focused on understanding the mechanisms of cWnt controlling their potential target genes in Ciona, such as the ParaHox genes (Gsx, Xlox and Cdx) that are the evolutionary sisters to the Hox genes, with roles in patterning the anterior-posterior axis in the central nervous system and gut. I had the opportunity to try both experiments with my two lab mates (Dr Nuria Torres-Aguila and Anastasia Ellis, see Figure 3) who use different ways to disrupt the cWnt signalling pathway.
Background
The cWnt pathway features the activation of the β-catenin transcription factor and modulation of specific target gene expression. T-cell factor/lymphoid enhancer (TCF/LEF) is the main transcription factor mediating the cWnt pathway. In the absence of Wnt ligands, responsive genes are generally repressed by TCF/LEF due to the transcriptional cofactor β-catenin constantly being degraded by the proteosome. When Wnt ligands interact with the transmembrane receptor proteins, several proteins (Frizzled, LRP5/6 and Disheveled) are brought to form a multimeric complex attached to the membrane, inhibiting the phosphorylation of β-catenin and its degradation. The stabilised β-catenin then converts the former repressor TCF/LEF into a transcriptional activator of the Wnt-target genes (Gilbert and Barresi, 2022).
Heat shock experiment
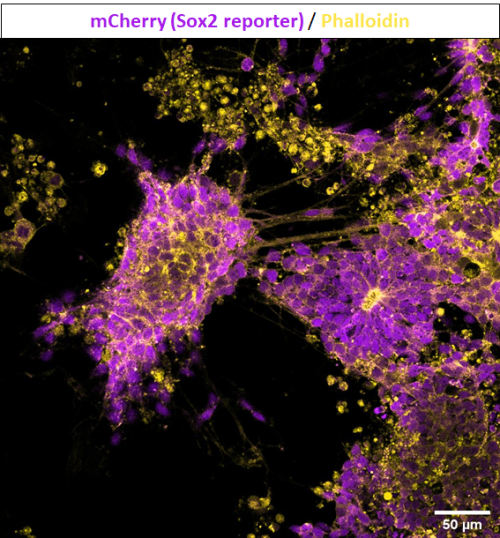
Controlling when and where TCF is expressed is useful to help analyse the function of TCF in cWnt signalling. Based on the heat-inducible cis-regulatory element initially characterized by Kawaguchi’s team (Kawaguchi et al., 2014), we tested the efficiency of a DNA construct with heat-inducible gene Ci-HSPA1/2/6-like and mCherry gene in Ciona embryogenesis to adapt this versatile technique to our species and population (i.e. a temperate Scottish population versus a more tropical Japanese population). Heat shock was initiated at different stages of development with different lengths of time and temperatures before being observed under an epifluorescence microscope.
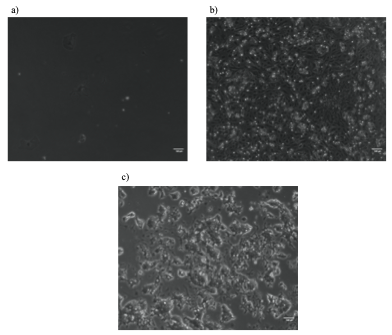
Although the precise heat-shock conditions need to be further refined, it was encouraging to see the induction of mCherry in embryos with this technique (Figure 1), raising the prospects of using the heat-shock approach to over-express genes like TCF in the near future. In this experiment, the microscope was probably the most exciting but also challenging piece of equipment, the microscope was probably the most exciting but also challenging piece of equipment I used. It took me some time to patiently check every detail like light intensity, exposure time, and magnification to ensure comparability between my images. The experience has been valuable for me to be familiar with this essential equipment for studying developmental biology.
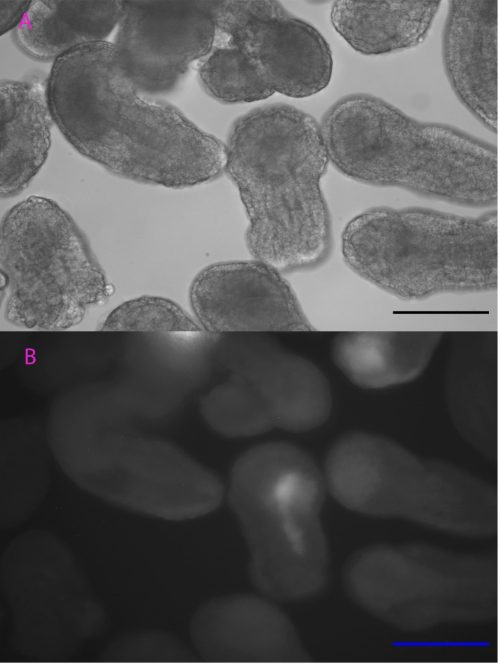
Scale bar: 100µm
Chemical treatment
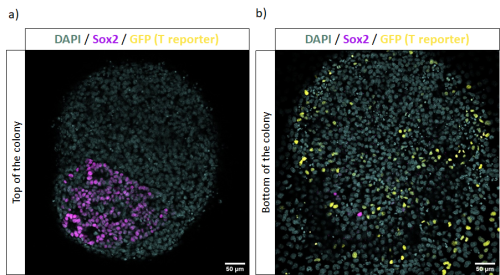
Chemical treatment investigated whether the TCF-dependent ParaHox gene Cdx is potentially directly controlled by the cWnt pathway. Pharmacological agents, iCRT-14 and azakenpaullone (Akp) were used to downregulate and upregulate the level of the cWnt pathway. Embryos were left to develop to the desired stage for recovery as a post treatment to distinguish rapid (potentially direct) responses from slower (possibly secondary) changes. The samples were processed for in-situ hybridization with a probe against the Cdx gene and images were captured by Nomarski microscopy.
The results showed that the expression of Cdx is possibly regulated by a secondary effect instead of directly disrupted by cWnt cascade, as the samples fixed immediately after treatment showed no obvious changes in expression (data not shown) whereas dramatic anterior extension of Cdx expression was seen with a recovery period in cWnt activator treatment (Figure 2). The lack of apparent response to iCRT14-treated samples may be due to TCF not normally being expressed in tail epidermal cells (Garstang et al., 2016). Further work aiming to check the Cdx expression in deeper cells where TCF is expressed will be conducted in the future.
All of this research helped me to appreciate the importance of time management in the lab. I had the opportunity to repeat the in-situ hybridization several times, and each time I got more skilled in the procedure. My first two times of in-situ hybridization turned out with either the embryos being accidentally lost because of I rushed during wash steps, or they were left in the solution for too long, so I stayed in the lab until very late. It is a relief that eventually they were successfully mounted to be observed, but wisely making use of time during short waits and multitasking largely improved the efficiency in my later experiments.
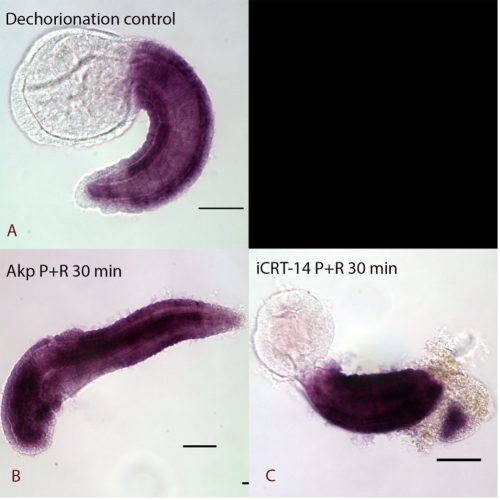
Expression of Cdx in Ciona intestinalis at late tailbud stage I A: Normal expression of Cdx at the late tailbud stage of dechorionated control samples. Expression is observed in the epidermis of the tail except at the very posterior end. B: 30 minutes pulse and recovery samples be treated by cWnt activator (Akp). It had dramatic anterior extension of Cdx expression and lost some of the anterior features. C: cWnt inhibitor (iCRT-14) pulse and recovery treated embryos with extension of Cdx expression to the posterior tip of the tail.
Pulse and immediately fixed samples did not show obvious changes in expression. (data not shown)
All the embryos are lateral views
scale bar: 50 µm
Personal experience
I appreciate that I have experienced the systematic research process as a whole. From collecting sea squirts in the harbour and manipulating the embryos, to finally assaying their responses to various treatments. I have tried many essential techniques in developmental biology and see how the experiments proceed. I learned how the protocols are designed and improved based on the techniques developed from previous research. Even finding the potential mistakes I made by recalling the steps with my supervisors was a valuable and rewarding experience. Thanks to Dave, Nuria, Anastasia and the friendly people around SOI, all of who created a warm and supportive environment. I am more comfortable working in the laboratory and more determined to pursue further studies now. Thanks to the BSDB and Gurdon scholarship for making this opportunity possible. It has been one of my best memories doing scientific exploration with such a great team, besides the beautiful East Sands beach at St. Andrews.

Reference list:
-Garstang M.G., Osborne P.W. and Ferrier D.E.K. (2016) TCF/Lef regulates the Gsx ParaHox gene in central nervous system development in chordates. BMC Evolutionary Biology 16:57.
-Gilbert S.F. and Baresi M.J (2022) Developmental Biology 12th edition Chapter 4 Cell to cell communication. Oxford University Press 109-111
-Kawaguchi A, Utsumi N, Morita M, Ohya A, Wada S. (2014) Application of the cis-regulatory region of a heat-shock protein 70 gene to heat-inducible gene expression in the ascidian Ciona intestinalis. Genesis. ;53(1):170-82.


 (4 votes)
(4 votes)

 (No Ratings Yet)
(No Ratings Yet)