August in preprints
Posted by the Node, on 1 September 2020
Welcome to our monthly trawl for developmental biology (and related) preprints.
The preprints this month are hosted on bioRxiv and arXiv – use these links to get to the section you want.
Developmental biology
| Stem cells, regeneration & disease modelling
Evo-devo & evo
Cell biology
Modelling
Tools & resources
Research practice & education
Developmental biology
| Patterning & signalling
Deciphering and modelling the TGF-β signalling interplays specifying the dorsal-ventral axis of the sea urchin embryo
Swann Floc’hlay, Maria Dolores Molina, Céline Hernandez, Emmanuel Haillot, Morgane Thomas-Chollier, Thierry Lepage, Denis Thieffry

The tolerance to hypoxia is defined by a time-sensitive response of the gene regulatory network in sea urchin embryos
Majed Layous, Lama Khalaily, Tsvia Gildor, Smadar Ben-Tabou de-Leon
Calcium-vesicles perform active diffusion in the sea urchin embryo during larval biomineralization
Mark R. Winter, Miri Morgulis, Tsvia Gildor, Andrew R. Cohen, Smadar Ben-Tabou de-Leon
Cellular pathways of calcium transport and concentration towards mineral formation in sea urchin larvae
Keren Kahil, Neta Varsano, Andrea Sorrentino, Eva Pereiro, Peter Rez, Steve Weiner, Lia Addadi
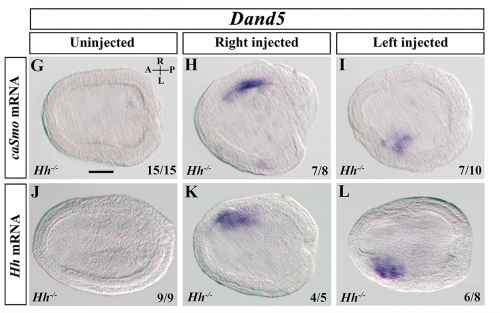
A transitory signaling center controls timing of primordial germ cell differentiation
Torsten U. Banisch, Maija Slaidina, Selena Gupta, Lilach Gilboa, Ruth Lehmann
Drosophila macrophage self-renewal is regulated by transient expression of PDGF- and VEGF-related factor 2
Daniel Bakopoulos, James C. Whisstock, Coral G. Warr, Travis K. Johnson
Drosophila Activin signaling promotes muscle growth through InR/dTORC1 dependent and independent processes
Myung-Jun Kim, Michael B. O’Connor
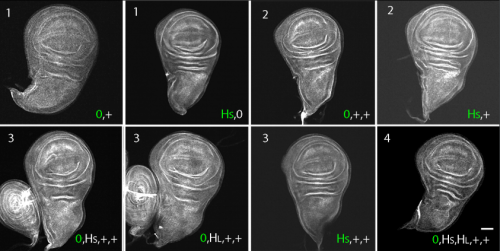
Regulated delivery controls Drosophila Hedgehog, Wingless and Decapentaplegic signaling
Ryo Hatori, Thomas B. Kornberg
CREB mediates the C. elegans dauer polyphenism through direct and cell-autonomous regulation of TGF-β expression
Jisoo Park, Hyekyung Oh, Do-Young Kim, YongJin Cheon, Yeon-Ji Park, Scott J. Neal, Abdul Rouf Dar, Rebecca A. Butcher, Piali Sengupta, Daewon Kim, Kyuhyung Kim
The N-Glycome regulates the endothelial-to-hematopoietic transition
Dionna M. Kasper, Jared Hintzen, Yinyu Wu, Joey J. Ghersi, Hanna K. Mandl, Kevin E. Salinas, William Armero, Zhiheng He, Ying Sheng, Yixuan Xie, Daniel W. Heindel, Eon Joo Park, William C. Sessa, Lara K. Mahal, Carlito Lebrilla, Karen K. Hirschi, Stefania Nicoli
Neural crest-derived neurons are replaced by a newly identified mesodermal lineage in the post-natal and aging enteric nervous system
Subhash Kulkarni, Monalee Saha, Laren S Becker, Zhuolun Wang, Guosheng Liu, Jenna Leser, Mithra Kumar, Shriya Bakhshi, Matthew J Anderson, Mark Lewandoski, Jared Slosberg, Sushma Nagaraj, Elizabeth Vincent, Loyal Goff, Pankaj Jay Pasricha
Patterning the embryonic pulmonary mesenchyme
Katharine Goodwin, Jacob M. Jaslove, Hirotaka Tao, Min Zhu, Sevan Hopyan, Celeste M. Nelson
Live imaging of avian embryos revealing a new head precursor map and the role for the anterior mesendoderm in brain development
Koya Yoshihi, Kagayaki Kato, Hideaki Iida, Machiko Teramoto, Akihito Kawamura, Yusaku Watanabe, Mitsuo Nunome, Mikiharu Nakano, Yoichi Matsuda, Yuki Sato, Hidenobu Mizuno, Takuji Iwasato, Yasuo Ishii, Hisato Kondoh
Endoderm Nitric Oxide Signals to Regulate Nascent Development of Cardiac Progenitors in Chicken Embryos
Devan H Shah, Sujoy K Biswas, Adrian M Martin, Simone Bianco, Wilfred F Denetclaw
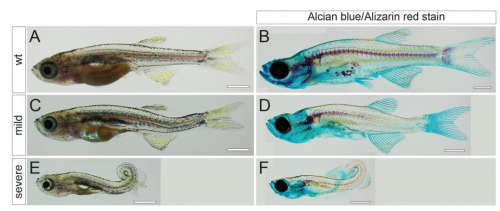
Postembryonic screen for mutations affecting spine development in zebrafish
Ryan S. Gray, Roberto Gonzalez, Sarah D. Ackerman, Ryoko Minowa, Johanna F. Griest, Melisa N. Bayrak, Benjamin Troutwine, Stephen Canter, Kelly R. Monk, Diane S. Sepich, Lilianna Solnica-Krezel
Fgf/Ets signalling in Xenopus ectoderm initiates neural induction and patterning in an autonomous and paracrine manners
Ikuko Hongo, Harumasa Okamoto
Rab7 mediates Wnt-dependent mesoderm patterning and gastrulation in Xenopus
Jennifer Kreis, Fee M. Wielath, Philipp Vick
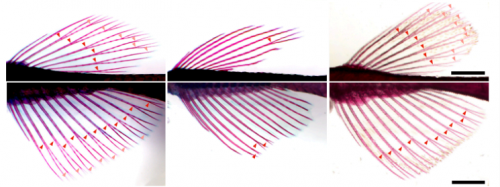
Thyroid hormone regulates proximodistal identity in the fin skeleton
Yinan Hu, Melody Harper, Benjamin Acosta, Joan Donahue, Hoa Bui, Hyungwoo Lee, Stacy Nguyen, Sarah McMenamin
Timed mesodermal FGF and BMP govern the multi-step thyroid specification
Benoit Haerlingen, Robert Opitz, Isabelle Vandernoot, Angelo Molinaro, Meghna Shankar, Pierre Gillotay, Achim Trubiroha, Sabine Costagliola
DYRK2 is a ciliary kinase involved 1 in vertebrate Hedgehog signal transduction
Nicholas Morante, Monika Abedin Sigg, Luke Strauskulage, David R. Raleigh, Jeremy F. Reiter
An evolutionarily conserved Lhx2-Ldb1 interaction regulates the acquisition of hippocampal cell fate and regional identity
Veena Kinare, Archana Iyer, Hari Padmanabhan, Geeta Godbole, Tooba Khan, Zeba Khatri, Upasana Maheshwari, Bhavana Muralidharan, Shubha Tole
Endothelial Cell Cycle State Determines Propensity for Arterial-Venous Fate
Nicholas W. Chavkin, Gael Genet, Mathilde Poulet, Nafiisha Genet, Corina Marziano, Hema Vasavada, Elizabeth A. Nelson, Anupreet Kour, Stephanie P. McDonnell, Mahalia Huba, Kenneth Walsh, Karen K. Hirschi
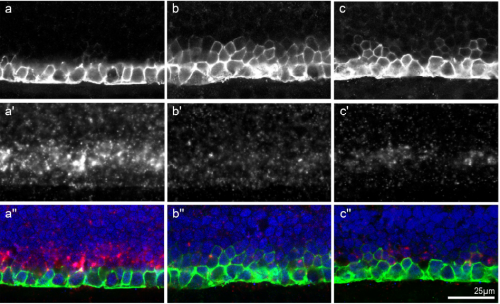
Sulf2a controls Shh-dependent neural fate specification in the developing spinal cord
Cathy Danesin, Romain Darche-Gabinaud, Nathalie Escalas, Vanessa Bouguetoch, Philippe Cochard, Amir Al Oustah, David Ohayon, Bruno Glise, Cathy Soula
A Highly Conserved Shh Enhancer Coordinates Hypothalamic and Craniofacial Development
Zoe Crane-Smith, Jeffrey Schoenebeck, Katy A Graham, Paul S Devenney, Lorraine Rose, Mark Ditzell, Eve Anderson, Joseph I Thomson, Natasha Klenin, Deborah M Kurrasch, Laura A Lettice, Robert E Hill
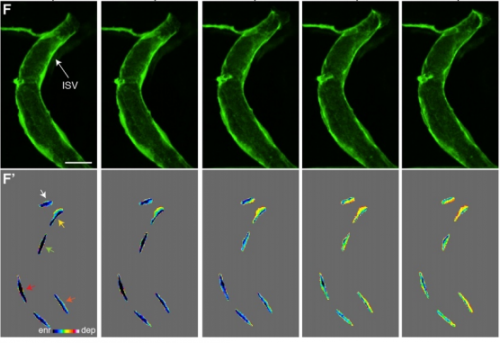
Live-imaging of endothelial Erk activity reveals dynamic and sequential signalling events during regenerative angiogenesis
Kazuhide S. Okuda, Mikaela Keyser, David B. Gurevich, Caterina Sturtzel, Scott Patterson, Huijun Chen, Mark Scott, Nicholas D. Condon, Paul Martin, Martin Distel, Benjamin M. Hogan
Midkine in chick and mouse retinas: neuroprotection, glial reactivity and the formation of Müller glia-derived progenitor cells
Warren A. Campbell, Amanda Fritsch-Kelleher, Isabella Palazzo, Thanh Hoang, Seth Blackshaw, Andy J. Fischer
The BMP antagonist Gremlin1 contributes to the development of cortical excitatory neurons, motor balance and fear responses
Mari Ichinose, Nobumi Suzuki, Tongtong Wang, Hiroki Kobayashi, Laura Vrbanac, Jia Q Ng, Josephine A Wright, Tamsin R M Lannagan, Krystyna A Gieniec, Martin Lewis, Ryota Ando, Atsushi Enomoto, Simon Koblar, Paul Thomas, Daniel L Worthley, Susan L Woods
A dynamic and spatially periodic micro-pattern of HES5 expression underlies the probability of neuronal differentiation in the mouse spinal cord
V Biga, J Hawley, E Johns, D Han, J Kursawe, P Glendinning, C.S Manning, N Papalopulu
Multifaceted functions of Rab23 on primary cilium and Hedgehog signaling-mediated granule cell proliferation
CHH Hor, WY Leong, ELK Goh
Maternal iron deficiency perturbs embryonic cardiovascular development
Jacinta I. Kalisch-Smith, Nikita Ved, Dorota Szumska, Jacob Munro, Michael Troup, Shelley E. Harris, Aimée Jacquemot, Jack J. Miller, Eleanor M. Stuart, Magda Wolna, Emily Hardman, Fabrice Prin, Eva Lana-Elola, Rifdat Aoidi, Elizabeth M. C. Fisher, Victor L. J. Tybulewicz, Timothy J. Mohun, Samira Lakhal-Littleton, Eleni Giannoulatou, Duncan B. Sparrow
The DCC receptor regulates astroglial development essential for telencephalic morphogenesis and corpus callosum formation
Laura Morcom, Ilan Gobius, Ashley P L Marsh, Rodrigo Suárez, Caitlin Bridges, Yunan Ye, Laura R Fenlon, Yvrick Zagar, Amelia M Douglass, Amber-Lee S Donahoo, Thomas Fothergill, Samreen Shaikh, Peter Kozulin, Timothy J Edwards, Helen M Cooper, IRC5 Consortium, Elliott H Sherr, Alain Chédotal, Richard J Leventer, Paul J Lockhart, Linda J Richards
The SWELL1-LRRC8 complex regulates endothelial AKT-eNOS-mTOR signaling and vascular function
Ahmad F. Alghanem, Chau Ta, Joshua M. Maurer, Susheel K. Gunasekar, Ashutosh Kumar, Urooj Fatima, Chen Kang, Litao Xie, Oluwaseun Adeola, Javier Abello, Megan Riker, Macaulay Elliot-Hudson, Rachel A. Minerath, Amber Stratman, Chad E. Grueter, Robert F. Mullins, Rajan Sah
The ESCRT protein CHMP5 restrains skeletal progenitor cell senescence by preserving endo-lysosomal-mitochondrial network
Xianpeng Ge, Lizhi He, Haibo Liu, Cole M. Haynes, Jae-Hyuck Shim
Specification of oxytocinergic and vasopressinergic circuits in the developing mouse brain
M. Pilar Madrigal Verdú, Sandra Jurado
Purinergic signaling controls spontaneous activity in the auditory system throughout early development
Travis A. Babola, Sally Li, Zhirong Wang, Calvin Kersbergen, Ana Belén Elgoyhen, Thomas Coate, Dwight Bergles
Efferent feedback enforces bilateral coupling of spontaneous activity in the developing auditory system
Yixiang Wang, Maya Sanghvi, Alexandra Gribizis, Yueyi Zhang, Lei Song, Barbara Morley, Daniel G. Barson, Joseph Santos-Sacchi, Dhasakumar Navaratnam, Michael Crair
Early enforcement of cell identity by a functional component of the terminally differentiated state
Zahra Bahrami-Nejad, Tinghuan Chen, Stefan Tholen, Zhi-Bo Zhang, Atefeh Rabiee, Michael L. Zhao, Fredric B. Kraemer, Mary N. Teruel
Oviduct epithelial cells constitute two developmentally distinct lineages that are spatially separated along the distal-proximal axis
Matthew J Ford, Keerthana Harwalkar, Alain S Pacis, Helen Maunsell, Yu Chang Wang, Dunarel Badescu, Katie Teng, Nobuko Yamanaka, Maxime Bouchard, Jiannis Ragoussis, Yojiro Yamanaka
Prom1 expression does not mark a stem/progenitor population in the mouse oviduct epithelium
Matthew J Ford, Yojiro Yamanaka
Wt1-expressing cells contribute to mesoderm-derived tissues in intestine and mesentery in two distinct phases during murine embryonic development
Suad Alghamdi, Thomas P Wilm, Shanthi Beglinger, Michael Boyes, Helen Tanton, Fiona Mutter, Joanna Allardyce, Veronica Foisor, Ben Middlehurst, Lauren Carr, Kelly Ward, Tarek Benameur, Thomas Butts, Nicholas Hastie, Bettina Wilm
| Morphogenesis & mechanics
Cell surface fluctuations regulate early embryonic lineage sorting
Ayaka Yanagida, Christopher Revell, Giuliano G. Stirparo, Elena Corujo-Simon, Irene M. Aspalter, Ruby Peters, Henry De Belly, Davide A. D. Cassani, Sarra Achouri, Raphael Blumenfeld, Kristian Franze, Ewa K. Paluch, Jennifer Nichols, Kevin J. Chalut
Symmetry breaking and de-novo axis formation in hydra spheroids: the microtubule cytoskeleton as a pivotal element
Heike Sander, Aravind Pasula, Mathias Sander, Varun Giri, Emmanuel Terriac, Franziska Lautenschlaeger, Albrecht Ott
The microtubule organization in the zebrafish yolk adapts to transgene-mediated phenotypic variations
Maria Marsal, Matteo Bernardello, Emilio J. Gualda, Pablo Loza-Alvarez
Which actin genes are necessary for zebrafish heart development and function?
Kendal Prill, Matiyo Ojehomon, Love Sandhu, Suchandrima Dutta, John F. Dawson
Morphogenesis of the islets of Langerhans is guided by extra-endocrine Slit2/3 signals
Jennifer M. Gilbert, Melissa T. Adams, Nadav Sharon, Hariharan Jayaraaman, Barak Blum
Mechanical feedback and robustness of apical constrictions in Drosophila embryo ventral furrow formation
Michael C. Holcomb, Guo-Jie Jason Gao, Mahsa Servati, Dylan Schneider, Presley K. McNeely, Jeffrey H. Thomas, Jerzy Blawzdziewicz
Adhesion-mediated heterogeneous actin organization governs apoptotic cell extrusion
Anh Phuong Le, Jean-François Rupprecht, René-Marc Mège, Yusuke Toyama, Chwee Teck Lim, Benoît Ladoux
Cell-Cell Adhesion During Nephron Development Is Driven by Wnt/PCP Formin Daam1
Vanja Krneta-Stankic, Mark Corkins, Adriana Paulucci-Holthauzen, Malgorzata Kloc, Andrew Gladden, Rachel Miller
The cytoskeleton adaptor protein Sorbs1 controls the development of lymphatic and venous vessels in zebrafish
Alexandra Veloso, Anouk Bleuart, Tanguy Orban, Jonathan Bruyr, Pauline Cabochette, Raoul F.V. Germano, Alice Bernard, Benoit Vanhollebeke, Maud Martin, Franck Dequiedt
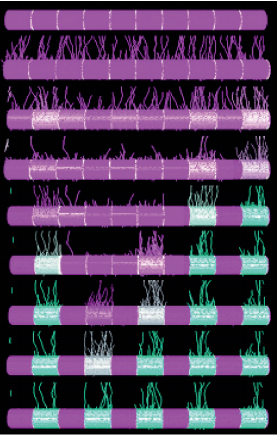
Active Perception during Angiogenesis: Filopodia speed up Notch selection of tip cells in silico and in vivo
Bahti Zakirov, Georgios Charalambous, Irene M. Aspalter, Kelvin Van-Vuuren, Thomas Mead, Kyle Harrington, Raphael Thuret, Erzsébet Ravasz Regan, Shane Paul Herbert, Katie Bentley

GATA4 regulates epithelial morphogenesis in the developing mouse stomach to promote establishment of a glandular columnar epithelium
Ann DeLaForest, Bridget M. Kohlnhofer, Olivia D. Franklin, Roman Stavniichuk, Cayla A. Thompson, Kirthi Pulakanti, Sridhar Rao, Michele A. Battle
Somite deformations buffer imprecise segment lengths to ensure left-right symmetry
Sundar R. Naganathan, Marko Popović, Andrew C. Oates
Cell intercalation driven by SMAD3 underlies secondary neural tube formation
Elena Gonzalez-Gobartt, José Blanco-Ameijeiras, Susana Usieto, Guillaume Allio, Bertrand Benazeraf, Elisa Martí
Twisting of the heart tube during cardiac looping is a tbx5-dependent and tissue-intrinsic process
Federico Tessadori, Fabian Kruse, Susanne C. van den Brink, Malou van den Boogaard, Vincent M. Christoffels, Jeroen Bakkers
Microglial trogocytosis and the complement system regulate axonal pruning in vivo
Tony K.Y. Lim, Edward S. Ruthazer
Epithelial layer unjamming shifts energy metabolism toward glycolysis
Stephen J. DeCamp, Victor M.K. Tsuda, Jacopo Ferruzzi, Stephan A. Koehler, John T. Giblin, Darren Roblyer, Muhammad H. Zaman, Scott T. Weiss, Margherita DeMarzio, Chan Young Park, Nicolas Chiu Ogassavara, Jennifer Mitchel, James P. Butler, Jeffrey J. Fredberg
Control of dynamic cell behaviors during angiogenesis and anastomosis by Rasip 1
Minkyoung Lee, Charles Betz, Ilkka Paatero, Niels Schellinx, Jianmin Yin, Christopher William Wilson, Weilan Ye, Markus Affolter, Heinz-Georg Belting
Marcksl1 modulates endothelial cell mechanoresponse to haemodynamic forces to control blood vessel shape and size
Igor Kondrychyn, Douglas J Kelly, Nuria Taberner, Akane Nomori, Kagayaki Kato, Jeronica Chong, Hiroyuki Nakajima, Satoru Okuda, Naoki Mochizuki, Li-Kun Phng
Planar polarization of cilia in the zebrafish floor-plate involves Par3-mediated posterior localization of highly motile basal bodies
Antoine Donati, Sylvie Schneider-Maunoury, Christine Vesque
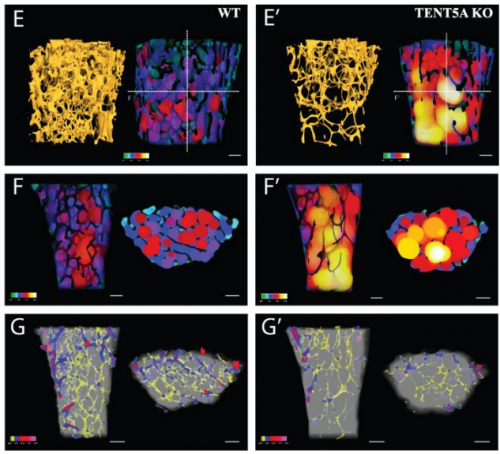
Cytoplasmic polyadenylation by TENT5A is required for proper bone formation
Olga Gewartowska, Goretti Aranaz Novaliches, Paweł S Krawczyk, Seweryn Mroczek, Monika Kusio-Kobiałka, Bartosz Tarkowski, Frantisek Spoutil, Oldrich Benada, Olga Kofroňová, Piotr Szwedziak, Dominik Cysewski, Jakub Gruchota, Marcin Szpila, Aleksander Chlebowski, Radislav Sedlacek, Jan Prochazka, Andrzej Dziembowski
Paranode stability requires UNC5B expression by oligodendrocytes
Omar de Faria Jr., Diane S. Nakamura, Samuel Clemot, Doyeun Kim, Mihai Victor Mocanu, Roland Pilgram, Jenea M. Bin, Edwin W. Wong, Amir Shmuel, Abbas Sadikot, Susan L. Ackerman, Timothy E. Kennedy
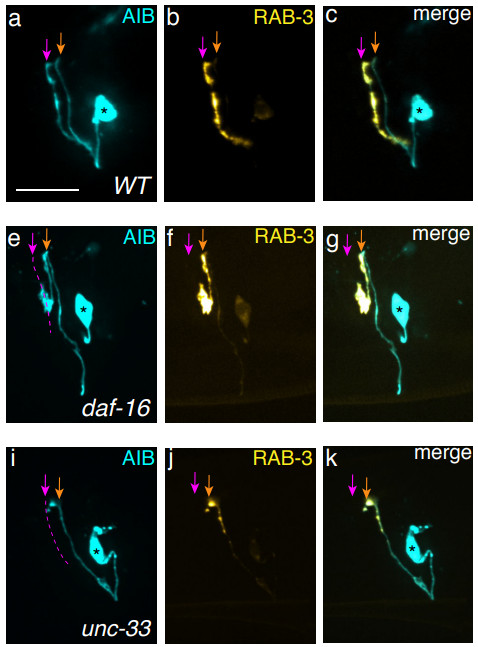
A neurite-zippering mechanism, mediated by layer-specific expression of IgCAMs, regulates synaptic laminar specificity in the C. elegans nerve ring neuropil
Titas Sengupta, Noelle L. Koonce, Mark W. Moyle, Leighton H. Duncan, Nabor Vázquez-Martínez, Xiaofei Han, Lin Shao, Yicong Wu, Anthony Santella, Li Fan, Zhirong Bao, William A. Mohler, Hari Shroff, Daniel A. Colón-Ramos
| Genes & genomes
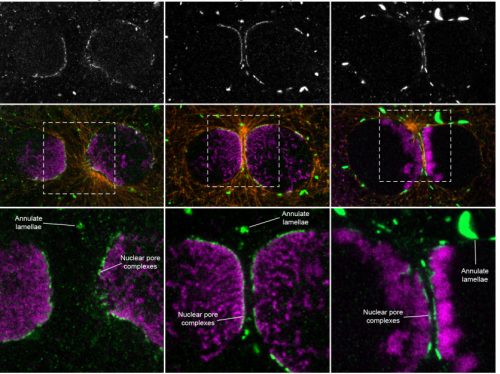
Parental genome unification is highly erroneous in mammalian embryos
Tommaso Cavazza, Antonio Z Politi, Patrick Aldag, Clara Baker, Kay Elder, Martyn Blayney, Andrea Lucas-Hahn, Heiner Niemann, Melina Schuh
Specification and epigenetic resetting of the pig germline exhibit conservation with the human lineage
Qifan Zhu, Fei Sang, Sarah Withey, Walfred Tang, Sabine Dietmann, Doris Klisch, Priscila Ramos-Ibeas, Haixin Zhang, Cristina E. Requena, Petra Hajkova, Matt Loose, M. Azim Surani, Ramiro Alberio
Human Embryonic Expression Identifies Novel Essential Gene Candidates
Monica Penon-Portmann, Jiyoo Chang, David R. Blair, Beatriz Rodriguez-Alonso, Hakan Cakmak, Aleksandar Rajkovic, Joseph T. Shieh
A human-specific structural variation at the ZNF558 locus controls a gene regulatory network during forebrain development
Pia A. Johansson, Per Ludvik Brattås, Christopher H. Douse, PingHsun Hsieh, Julien Pontis, Daniela Grassi, Raquel Garza, Marie E. Jönsson, Diahann A. M. Atacho, Karolina Pircs, Feride Eren, Yogita Sharma, Jenny Johansson, Didier Trono, Evan E. Eichler, Johan Jakobsson
Temporally-Divergent Regulatory Mechanisms Govern Neuronal Development and Diversification in the Neocortex
Wen Yuan, Sai Ma, Juliana R. Brown, Kwanho Kim, Vanessa Murek, Lucia Trastulla, Alexander Meissner, Simona Lodato, Ashwin Shetty, Joshua Z. Levin, Jason D. Buenrostro, Michael J. Ziller, Paola Arlotta
Identification of 3’ UTR motifs required for mRNA localization to myelin sheaths in vivo
Katie M. Yergert, Rebecca O’Rouke, Jacob H. Hines, Bruce Appel
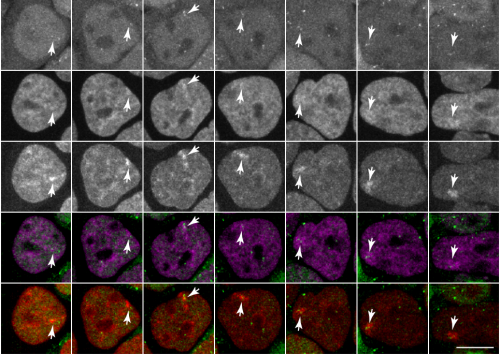
Tracking H3K27me3 and H4K20me1 dynamics during XCI reveals similarities in recruitment mechanism
Sjoerd J.D. Tjalsma, Mayako Hori, Yuko Sato, Aurelie Bousard, Akito Ohi, Ana Claudia Raposo, Julia Roensch, Agnes Le Saux, Jumpei Nogami, Kazumitsu Maehara, Tomoya Kujirai, Yasuyuki Ohkawa, Hitoshi Kurumizaka, Simao Teixeira da Rocha, Jan Jakub Zylicz, Hiroshi Kimura, Edith Heard
X-chromosome upregulation is dynamically linked to the X-inactivation state
Antonio Lentini, Christos Coucoravas, Nathanael Andrews, Martin Enge, Qiaolin Deng, Björn Reinius
Divergent DNA methylation signatures underlying X chromosome regulation in marsupials and eutherians
Devika Singh, Dan Sun, Andrew G. King, David E. Alquezar-Planas, Rebecca N. Johnson, David Alvarez-Ponce, Soojin V. Yi
Systematic investigation of imprinted gene expression and enrichment in the mouse brain explored at single-cell resolution
M. J. Higgs, M. J. Hill, R. M. John, A. R. Isles
Maternal factor PABPN1L is essential for maternal mRNA degradation during maternal-to-zygotic transition
Ying Wang, Tianhao Feng, Xiaodan Shi, Siyu Liu, Zerui Wang, Xin Zhang, Jintao Zhang, Shuqin Zhao, Junqiang Zhang, Xiufeng Ling, Mingxi Liu
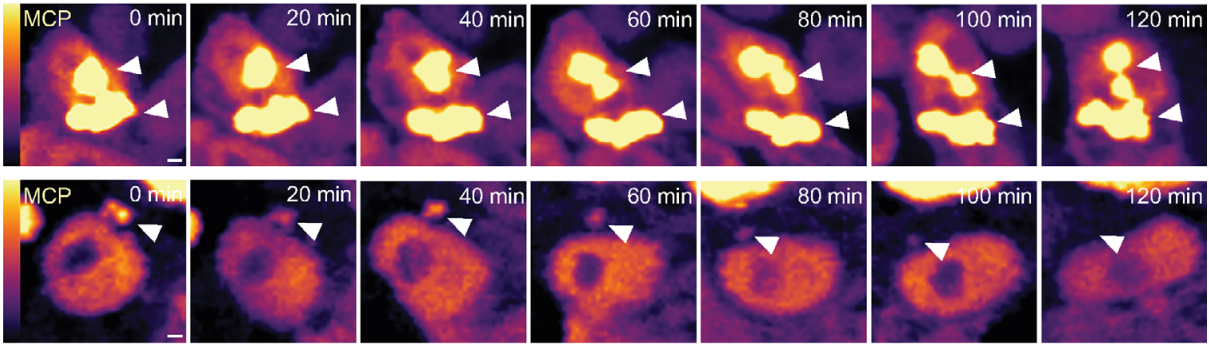
P-bodies are sites of rapid RNA decay during the neural crest epithelial—mesenchymal transition
Erica J. Hutchins, Michael L. Piacentino, Marianne E. Bronner
Transcriptome and epigenome characterization of mouse spermatogonial cells reveals distinct chromatin regulatory landscapes in postnatal and adult testis
Irina Lazar-Contes, Deepak K. Tanwar, Pierre-Luc Germain, Niharika Gaur, Isabelle M. Mansuy
Sequential onset and concurrent expression of miR-9 genomic loci in single cells contributes to the temporal increase of mature miR-9 in zebrafish neurogenesis
X. Soto, T. Minchington, R. Lea, J. Lee, N Papalopulu
Epigenetic change induced by in utero dietary challenge provokes phenotypic variability across multiple generations of mice
Mathew Van de Pette, Antonio Galvao, Steven J. Millership, Wilson To, Andrew Dimond, Chiara Prodani, Grainne McNamara, Ludovica Bruno, Alessandro Sardini, Zoe Webster, James McGinty, Paul French, Anthony G. Uren, Juan Castillo-Fernandez, Rosalind M. John, Anne C. Ferguson-Smith, Matthias Merkenschlager, Gavin Kelsey, Amanda G. Fisher
MiR-124 synergism with ELAVL3 enhances target gene expression to promote neuronal maturity
Ya-Lin Lu, Yangjian Liu, Matthew J. McCoy, Andrew S. Yoo
An early Sox2-dependent gene expression program required for hippocampal dentate gyrus development
Sara Mercurio, Chiara Alberti, Linda Serra, Simone Meneghini, Jessica Bertolini, Pietro Berico, Andrea Becchetti, Silvia K. Nicolis
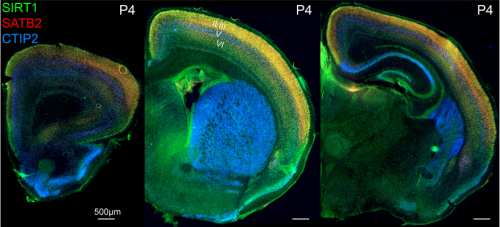
Fezf2 transient expression via modRNA with concurrent SIRT1 inhibition enhances differentiation of cortical subcerebral / corticospinal neuron identity from mES cells
Cameron Sadegh, Wataru Ebina, Anthony C. Arvanites, Lance S. Davidow, Lee L. Rubin, Jeffrey D. Macklis
Scaling of gene transcriptional gradients with brain size across mouse development
Hoi Yan Gladys Lau, Alex Fornito, Ben D. Fulcher
Gene regulatory networks controlling differentiation, survival, and diversification of hypothalamic Lhx6-expressing GABAergic neurons.
Dong Won Kim, Kai Liu, Zoe Qianyi Wang, Yi Stephanie Zhang, Abhijith Bathini, Matthew P. Brown, Sonia Hao Lin, Parris W. Washington, Changyu Sun, Susan Lindtner, Bora Lee, Hong Wang, Tomomi Shimogori, John L.R. Rubenstein, Seth Blackshaw
Orphan CpG islands boost the regulatory activity of poised enhancers and dictate the responsiveness of their target genes
Tomas Pachano, Victor Sanchez-Gaya, Maria Mariner-Fauli, Thais Ealo, Helena G Asenjo, Patricia Respuela, Sara Cruz-Molina, Wilfred van IJcken, David Landeira, Alvaro Rada-Iglesias
Single cell transcriptomics reveals lineage trajectory of the retinal ganglion cells in wild-type and Atoh7-null retinas
Fuguo Wu, Jonathan E. Bard, Julien Kann, Donald Yergeau, Darshan Sapkota, Yichen Ge, Zihua Hu, Jie Wang, Tao Liu, Xiuqian Mu
Single-cell analyses of the corneal epithelium: Unique cell types and gene expression profiles
Surabhi Sonam, Sushant Bangru, Kimberly J. Perry, Auinash Kalsotra, Jonathan J. Henry
Combined analysis of single cell RNA-Seq and ATAC-Seq data reveals putative regulatory toggles operating in native and iPS-derived retina
Anouk Georges, Haruko Takeda, Arnaud Lavergne, Michiko Mandai, Fanny Lepiemme, Latifa Karim, Loic Demeulenaere, Michael Schyns, Laurent Nguyen, Jean-Marie Rakic, Masayo Takahashi, Michel Georges
Cell-type, single-cell, and spatial signatures of brain-region specific splicing in postnatal development
Anoushka Joglekar, Andrey Prjibelski, Ahmed Mahfouz, Paul Collier, Susan Lin, Anna Katharina Schlusche, Jordan Marrocco, Stephen R. Williams, Bettina Haase, Ashley Hayes, Jennifer G. Chew, Neil I Weisenfeld, Man Ying Wong, Alexander N. Stein, Simon Hardwick, Toby Hunt, Zachary Bent, Olivier Fedrigo, Steven A. Sloan, Davide Risso, Erich D. Jarvis, Paul Flicek, Wenjie Luo, Geoffrey S. Pitt, Adam Frankish, August B. Smit, M. Elizabeth Ross, Hagen U. Tilgner
Clump sequencing exposes the spatial expression programs of intestinal secretory cells
Rita Manco, Inna Averbukh, Ziv Porat, Keren Bahar Halpern, Ido Amit, Shalev Itzkovitz
Quantitative lineage analysis identifies a long-term progenitor niche for the hepato-pancreato-biliary organ system
David Willnow, Uwe Benary, Anca Margineanu, Maria Lillina Vignola, Igor M. Pongrac, Zahra Karimaddini, Alessandra Vigilante, Jana Wolf, Francesca M. Spagnoli
SNPC-1.3 is a sex-specific transcription factor that drives male piRNA expression in C. elegans
Charlotte P. Choi, Rebecca J. Tay, Margaret R. Starostik, Suhua Feng, James J. Moresco, Brooke E. Montgomery, Emily Xu, Maya A. Hammonds, Michael C. Schatz, Taiowa A. Montgomery, John R. Yates III, Steven E. Jacobsen, John K. Kim
KH domain containing RNA-binding proteins coordinate with microRNAs to regulate Caenorhabditis elegans development
D Haskell, A Zinovyeva
Characterization of histone inheritance patterns in the Drosophila female germline
Elizabeth W. Kahney, Lydia Sohn, Kayla Viets-Layng, Robert Johnston, Xin Chen
Asymmetric histone inheritance regulates stem cell fate in Drosophila midgut
Emily Zion, Xin Chen
The molecular landscape of neural differentiation in the developing Drosophila brain revealed by targeted scRNA-seq and a multi-informatic analysis paradigm
Nigel S. Michki, Ye Li, Kayvon Sanjasaz, Yimeng Zhao, Fred Y. Shen, Logan A. Walker, Cheng-Yu Lee, Dawen Cai
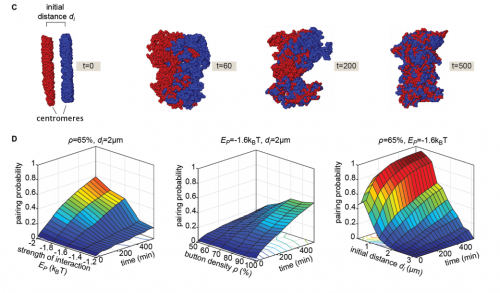
Live imaging and biophysical modeling support a button-based mechanism of somatic homolog pairing in Drosophila
Myron Barber Child VI, Jack R. Bateman, Amir Jahangiri, Armando Reimer, Nicholas C. Lammers, Nica Sabouni, Diego Villamarin, Grace C. McKenzie-Smith, Justine E. Johnson, Daniel Jost, Hernan G. Garcia
Quantitative imaging of RNA polymerase II activity in plants reveals the single-cell basis of tissue-wide transcriptional dynamics
Simon Alamos, Armando Reimer, Krishna K. Niyogi, Hernan G Garcia
Kinetic sculpting of the seven stripes of the Drosophila even-skipped gene
Augusto Berrocal, Nicholas Lammers, Hernan G. Garcia, Michael B. Eisen
The chromatin remodeler ISWI acts during Drosophila development to regulate adult sleep
Naihua N. Gong, Leela Chakravarti Dilley, Charlette E. Williams, Emilia H. Moscato, Milan Szuperak, Qin Wang, Matthew Jensen, Santhosh Girirajan, Tiong Yang Tan, Matthew A. Deardorff, Dong Li, Yuanquan Song, Matthew S. Kayser
Evolved differences in cis and trans regulation between the maternal and zygotic mRNA complements in the Drosophila embryo
Emily L. Cartwright, Susan E. Lott
An unexpected bifurcation in the Pointed transcriptional effector network contributes specificity and robustness to retinal cell fate acquisition
Chudong Wu, Jean-François Boisclair Lachance, Michael Z Ludwig, Ilaria Rebay
The 3’UTR of the orb2 gene encoding the Drosophila CPEB translation factor plays a critical role in spermatogenesis
Rudolf A. Gilmutdinov, Eugene N. Kozlov, Ludmila V. Olenina, Alexei A. Kotov, Justinn Barr, Mariya V. Zhukova, Paul Schedl, Yulii Shidlovskii
A critical role for Musashi in photoreceptor morphogenesis and Vision
Jesse Sundar, Fatimah Matalkah, Bohye Jeong, Peter Stoilov, Visvanathan Ramamurthy
Musashi regulates follicle stem cell maintenance and epithelial niche homeostasis in the Drosophila ovary
Nicole A. Siddall, Franca Casagranda, Nicole Dominado, James Heaney, Jessie M. Sutherland, Eileen A. McLaughlin, Gary R. Hime
| Stem cells, regeneration & disease modelling
Non-autonomous regulation of germline stem cell proliferation by somatic MPK-1/MAPK activity in C. elegans
Sarah Robinson-Thiewes, Benjamin Dufour, Pier-Olivier Martel, Xavier Lechasseur, Amani Ange Danielle Brou, Vincent Roy, Yunqing Chen, Judith Kimble, Patrick Narbonne
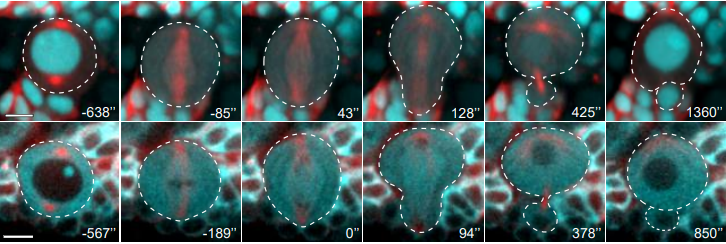
Asymmetric nuclear division of neural stem cells contributes to the formation of sibling nuclei with different identities
Chantal Roubinet, Ian J. White, Buzz Baum
Cystic proliferation of embryonic germ stem cells is necessary to reproductive success and normal mating behavior in medaka
Luisa F Arias Padilla, Diana C Castañeda-Cortés, Ivana F. Rosa, Rafael H Nóbrega, Juan I Fernandino
mRNA translation control by Dhx36 binding to 5′ UTR G-quadruplex structures is essential for skeletal muscle stem cell regenerative functions
Xiaona Chen, Jie Yuan, Guang Xue, Silvia Campanario Sanz, Di Wang, Wen Wang, Xi Mou, Mubarak Ishaq Ishaq Umar, Joan Isern, Yu Zhao, Liangqiang He, Yuying Li, Christopher J. Mann, Xiaohua Yu, Lei Wang, Eusebio Perdiguero, Wei Chen, Yuanchao Xue, Yoshikuni Nagamine, Chun-Kit Kwok, Hao Sun, Pura Muñoz-Cánoves, Huating Wang
Repopulating Kupffer Cells Originate Directly from Hematopoietic Stem Cells
Xu Fan, Pei Lu, Xianghua Cui, Peng Wu, Weiran Lin, Dong Zhang, Shongzong Yuan, Bing Liu, Fangyan Chen, Hong You, Handong Wei, Fuchu He, Jidong Jia, Ying Jiang
Induction of Ventral Spinal V0 Interneurons from Mouse Embryonic Stem Cells
Jennifer Pardieck, Manwal Harb, Shelly Sakiyama-Elbert
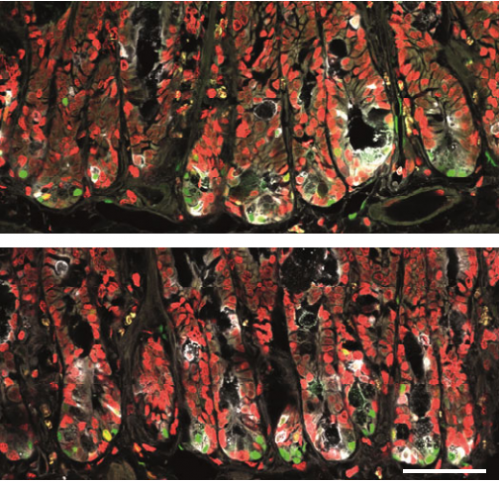
Wnt/PCP-primed intestinal stem cells directly differentiate into enteroendocrine or Paneth cells
Anika Böttcher, Maren Büttner, Sophie Tritschler, Michael Sterr, Alexandra Aliluev, Lena Oppenländer, Ingo Burtscher, Steffen Sass, Martin Irmler, Johannes Beckers, Christoph Ziegenhain, Wolfgang Enard, Andrea C. Schamberger, Fien M. Verhamme, Oliver Eickelberg, Fabian J. Theis, Heiko Lickert
Human muscle stem cells are refractory to aging
James S. Novak, Davi A.G. Mázala, Marie Nearing, Nayab F. Habib, Tessa Dickson, Olga B. Ioffe, Brent T. Harris, Marie N. Fidelia-Lambert, Christopher T. Rossi, D. Ashely Hill, Kathryn R. Wagner, Eric P. Hoffman, Terence A. Partridge
A comprehensive transcriptome signature of murine hematopoietic stem cell aging
Arthur Flohr Svendsen, Daozheng Yang, Seka Lazare, Erik Zwart, Albertina Ausema, Gerald de Haan, Leonid V. Bystrykh
Age-independent influence of hematopoietic stem and progenitor cell populations during hematopoietic reconstitution
Frauke Gotzhein, Tim Aranyossy, Lars Thielecke, Tanja Sonntag, Vanessa Thaden, Boris Fehse, Ingo Mueller, Ingmar Glauche, Kerstin Cornils
Developmental stage-specific changes in protein synthesis differentially sensitize hematopoietic stem cells and erythroid progenitors to impaired ribosome biogenesis
Jeffrey A. Magee, Robert A.J. Signer
Impaired Fetal Lung Development can be Rescued by Administration of Extracellular Vesicles Derived from Amniotic Fluid Stem Cells
Lina Antounians, Vincenzo D. Catania, Louise Montalva, Benjamin D. Liu, Huayun Hou, Cadia Chan, Andreea C. Matei, Areti Tzanetakis, Bo Li, Rebeca Lopes Figueira, Karina Miura da Costa, Amy P. Wong, Robert Mitchell, Anna L. David, Ketan Patel, Paolo De Coppi, Lourenço Sbragia Neto, Michael D. Wilson, Janet Rossant, Augusto Zani
Myogenin is an Essential Regulator of Adult Myofibre Growth and Muscle Stem Cell Homeostasis
Massimo Ganassi, Sara Badodi, Kees Wanders, Peter S. Zammit, Simon M. Hughes
Macrophages provide a transient muscle stem cell niche via NAMPT secretion
Dhanushika Ratnayake, Phong D. Nguyen, Fernando J. Rossello, Verena C. Wimmer, Abdulsalam I. Isiaku, Laura A. Galvis, Alasdair J. Wood, Ziad Julier, Thomas Boudier, Viola Oorschot, Kelly L. Rogers, Mikaël M. Martino, Christophe Marcelle, Graham J. Lieschke, Jeroen Bakkers, Peter D. Currie
Myofiber stretch induces tensile and shear deformation of muscle stem cells in their native niche
Mohammad Haroon, Jenneke Klein-Nulend, Astrid D. Bakker, Jianfeng Jin, Carla Offringa, Fabien Le Grand, Lorenzo Giordani, Karen J. Liu, Robert D. Knight, Richard T. Jaspers
SDC3 acts as a timekeeper of myogenic differentiation by regulating the insulin/AKT/mTOR axis in muscle stem cell progeny
Fiona K. Jones, Alexander Phillips, Andrew R. Jones, Addolorata Pisconti
Single-cell analysis identifies TCF4 and ID3 as a molecular switch of mammary epithelial stem cell differentiation
Koon-Kiu Yan, Erin Nekritz, Bensheng Ju, Xinran Dong, Rachel Werner, Dayanira Alsina-Beauchamp, Celeste Rosencrance, Partha Mukhopadhyay, Qingfei Pan, Andrej Gorbatenko, Liang Ding, Yanyan Wang, Chenxi Qian, Hao Shi, Bridget Shaner, Sivaraman Natarajan, Hongbo Chi, John Easton, Jose Silva, Jiyang Yu
Using single-cell entropy to describe the dynamics of reprogramming and differentiation of induced pluripotent stem cells
Yusong Ye, Zhuoqin Yang, Meixia Zhu, Jinzhi Lei
PU.1 drives specification of pluripotent stem cell-derived endothelial cells to LSEC-like cells
Jonathan De Smedt, Elise Anne van Os, Irene Talon, Sreya Ghosh, Burak Toprakhisar, Rodrigo Furtado Madeiro Da Costa, Samantha Zaunz, Marta Aguirre Vazquez, Ruben Boon, Pieter Baatsen, Ayla Smout, Stefaan Verhulst, Leo A. van Grunsven, Catherine M. Verfaillie
Histone demethylase complexes KDM3A and KDM3B cooperate with OCT4/SOX2 to construct pluripotency gene regulatory network
Zhenshuo Zhu, Xiaolong Wu, Qun Li, Juqing Zhang, Shuai Yu, Qiaoyan Shen, Zhe Zhou, Qin Pan, Wei Yue, Dezhe Qin, Ying Zhang, Wenxu Zhao, Rui Zhang, Sha Peng, Na Li, Shiqiang Zhang, Anmin Lei, Yi-Liang Miao, Zhonghua Liu, Xingqi Chen, Huayan Wang, Mingzhi Liao, Jinlian Hua
Single cell RNA sequence analysis of human bone marrow samples reveals new targets for isolation of skeletal stem cells using DNA-coated gold nanoparticles
Elloise Matthews, Stuart Lanham, Kate White, Maria-Eleni Kyriazi, Konstantina Alexaki, Afaf H. El-Sagheer, Tom Brown, Antonios G. Kanaras, Jonathan West, Ben D. MacArthur, Patrick S. Stumpf, Richard O.C. Oreffo
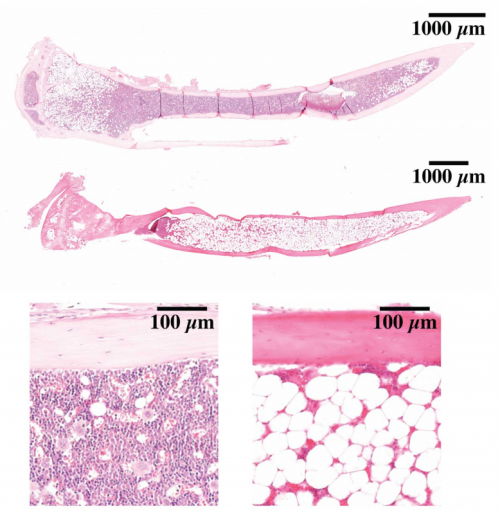
Hematopoietic stem cells fail to regenerate following inflammatory challenge
Ruzhica Bogeska, Paul Kaschutnig, Malak Fawaz, Ana-Matea Mikecin, Marleen Büchler-Schäff, Stella Paffenholz, Noboru Asada, Felix Frauhammer, Florian Buettner, Melanie Ball, Julia Knoch, Sina Stäble, Dagmar Walter, Amelie Petri, Martha J. Carreño-Gonzalez, Vinona Wagner, Benedikt Brors, Simon Haas, Daniel B. Lipka, Marieke A.G. Essers, Tim Holland-Letz, Jan-Philipp Mallm, Karsten Rippe, Paul S. Frenette, Michael A. Rieger, Michael D. Milsom
Coordinated interactions between endothelial cells and macrophages in the islet microenvironment promote beta cell regeneration
Diane C Saunders, Kristie I Aamodt, Tiffany M Richardson, Alec Hopkirk, Zoya Khan, Radhika Aramandla, Greg Poffenberger, Regina Jenkins, David K Flaherty, Nripesh Prasad, Shawn Levy, Alvin Powers, Marcela Brissova
Dendrimer-targeted immunosuppression of microglia reactivity super-accelerates photoreceptor regeneration in the zebrafish retina
Kevin B. Emmerich, David T. White, Siva P. Kambhamptati, Grace Y. Lee, Tian-Ming Fu, Arpan Sahoo, Meera T. Saxena, Eric Betzig, Rangaramanujam M. Kannan, Jeff S. Mumm
Time-resolved expression analysis comparing two selective retinal cell ablation paradigms in zebrafish reveals shared and cell-specific regenerative regulatory networks.
Steven L Walker, Guohua Wang, Kevin B Emmerich, Fang Wang, David T White, Meera T Saxena, Yong Teng, Jiang Qian, Jeff S Mumm
Non-canonical Hedgehog signaling regulates spinal cord and muscle regeneration
Laura N Borodinsky, Andrew M Hamilton
Impairment of the Hif-1α regulatory pathway in Foxn1-deficient (Foxn1-/-) mice affects the skin wound healing process
Sylwia Machcinska, Marta Kopcewicz, Joanna Bukowska, Katarzyna Walendzik, Barbara Gawronska-Kozak
Osmolarity-independent electrical cues guide rapid response to injury in zebrafish epidermis
Andrew S. Kennard, Julie A. Theriot
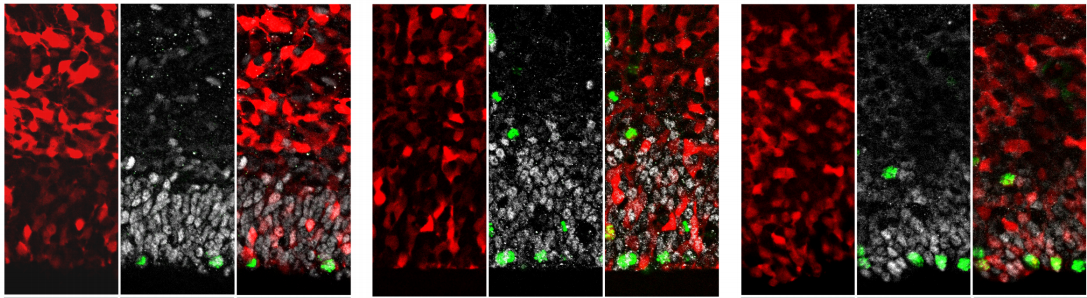
Endosomal trafficking defects alter neural progenitor proliferation and cause microcephaly
Jacopo A. Carpentieri, Amandine Di Cicco, David Andreau, Laurence Del Maestro, Fatima El Marjou, Laure Coquand, Jean-Baptiste Brault, Nadia Bahi-Buisson, Alexandre D. Baffet
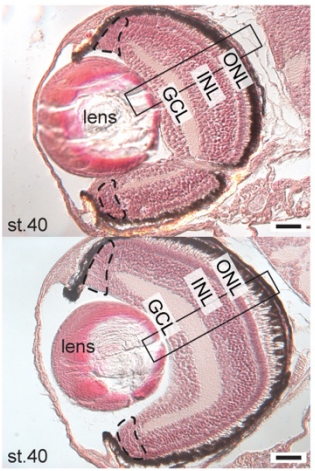
The medaka alg2 mutant is a model for hypo-N-glycosylation-associated retinitis pigmentosa
Sevinç Gücüm, Roman Sakson, Marcus Hoffmann, Valerian Grote, Lars Beedgen, Christian Thiel, Erdmann Rapp, Thomas Ruppert, Joachim Wittbrodt, Thomas Thumberger
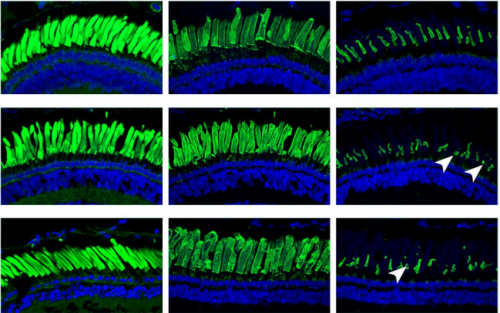
Genetically-modified X. laevis indicate distinct roles for prominin-1 and photoreceptor cadherin in outer segment morphogenesis and retinal dystrophy
Brittany J Carr, Paloma Stanar, Orson L Moritz
Identification of MYOM2 as a candidate gene in hypertrophic cardiomyopathy and Tetralogy of Fallot and its functional evaluation in the Drosophila heart
Emilie Auxerre-Plantié, Tanja Nielsen, Marcel Grunert, Olga Olejniczak, Andreas Perrot, Cemil Özcelik, Dennis Harries, Faramarz Matinmehr, Cristobal Dos Remedios, Christian Mühlfeld, Theresia Kraft, Rolf Bodmer, Georg Vogler, Silke R. Sperling
Modeling Genetic Epileptic Encephalopathies using Brain Organoids
Daniel J. Steinberg, Afifa Saleem, Srinivasa Rao Repudi, Ehud Banne, Muhammad Mahajnah, Jacob H. Hanna, Peter L. Carlen, Rami I. Aqeilan
Emergence of Non-Canonical Parvalbumin-Containing Interneurons in Hippocampus of a Murine Model of Type I Lissencephaly
Tyler G. Ekins, Vivek Mahadevan, Yajun Zhang, James A. D’Amour, Timothy Petros, Chris J. McBain
A Bbs5 mouse model reveals pituitary cilia contributions to developmental abnormalities
Melissa R. Bentley, Staci E. Engle, Courtney J. Haycraft, Reagan S. Andersen, Mandy J. Croyle, Kelsey R. Clearman, Addison B. Rains, Nicolas F. Berbari, Bradley K. Yoder
Role of matrix metalloproteinase-9 in neurodevelopmental disorders and experience-dependent plasticity in Xenopus tadpoles
Sayali Gore, Eric J. James, Lin-Chien Huang, Jenn J. Park, Andrea Berghella, Adrian Thompson, Hollis T. Cline, Carlos D. Aizenman
Extracellular matrix and cyclic stretch alter fetal cardiomyocyte proliferation and maturation in a rodent model of heart hypoplasia
Matthew C. Watson, Corin Williams, Raymond M. Wang, Luke R. Perreault, Kelly E. Sullivan, Whitney L. Stoppel, Lauren D. Black III
Identification of limb-specific Lmx1b auto-regulatory modules with Nail-Patella Syndrome pathogenicity
Endika Haro, Florence Petit, Charmaine U. Pira, Conor D. Spady, Lauren A. Ivey, Austin L. Gray, Fabienne Escande, Anne-Sophie Jourdain, Andy Nguyen, Florence Fellmann, Jean-Marc Good, Christine Francannet, Sylvie Manouvrier-Hanu, Marian A. Ros, Kerby C. Oberg
Targeted Correction and Functional Recovery in Achondroplasia Patient-Derived iPSCs
Huan Zou, Mingfeng Guan, Yundong Li, Fang Luo, Wenyuan Wang, Yiren Qin
Adipose tissue developmental growth constraints uncouple fat distribution from glucose metabolism in two mouse models of obesity
Zachary L. Sebo, Christopher Church, Ryan Berry, Matthew S. Rodeheffer
Zbtb16 regulates social cognitive behaviors and neocortical development
Noriyoshi Usui, Stefano Berto, Ami Konishi, Makoto Kondo, Genevieve Konopka, Hideo Matsuzaki, Shoichi Shimada
Ap2s1 mutation in mice causes familial hypocalciuric hypercalcemia type 3
Fadil M. Hannan, Mark Stevenson, Asha L. Bayliss, Victoria J. Stokes, Michelle Stewart, Kreepa G. Kooblall, Caroline M. Gorvin, Gemma Codner, Lydia Teboul, Sara Wells, Rajesh V. Thakker
Humanization of Drosophila Gαo to model GNAO1 paediatric encephalopathies
Mikhail Savitsky, Gonzalo P. Solis, Vladimir L. Katanaev
b3galt6 knock-out zebrafish recapitulate β3GalT6-deficiency disorders in human and reveal a trisaccharide proteoglycan linkage region
Sarah Delbaere, Adelbert De Clercq, Shuji Mizumoto, Fredrik Noborn, Jan Willem Bek, Lien Alluyn, Charlotte Gistelinck, Delfien Syx, Phil L. Salmon, Paul J. Coucke, Göran Larson, Yamada shuhei, Andy Willaert, Fransiska Malfait
Molecular markers characterization determining cell fate specification in an adult neurogenesis model of Alzheimer’s disease
Idoia Blanco-Luquin, Juan Cabello, Amaya Urdánoz-Casado, Blanca Acha, Eva Ma Gómez-Orte, Miren Roldan, Diego R. Pérez-Rodríguez, Maite Mendioroz
Molecular and electrophysiological features of spinocerebellar ataxia type seven in induced pluripotent stem cells
Richard J Burman, Lauren M Watson, Danielle C Smith, Joseph V Raimondo, Robea Ballo, Janine Scholefield, Sally A Cowley, Matthew JA Wood, Susan H Kidson, Leslie J Greenberg
| Plant development
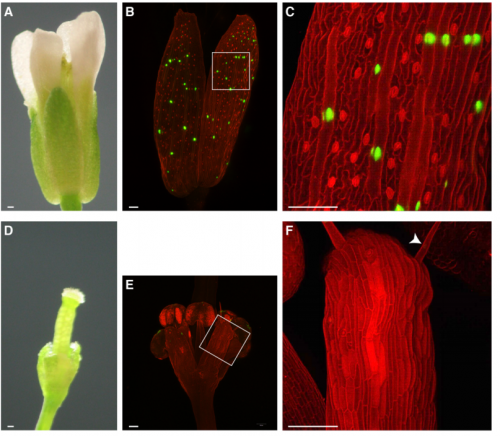
A giant cell enhancer achieves cell-type specificity through activation via TCP and repression by Dof transcription factors
Lilan Hong, Clint S. Ko, S. Earl Kang, Jose L. Pruneda-Paz, Adrienne H. K. Roeder
PIEZO ion channel is required for root mechanotransduction in Arabidopsis thaliana
Seyed A. R. Mousavi, Adrienne E Dubin, Wei-Zheng Zeng, Adam M. Coombs, Khai Do, Darian A. Ghadiri, Chennan Ge, Yunde Zhao, Ardem Patapoutian
Single nucleus analysis of Arabidopsis seeds reveals new cell types and imprinting dynamics
Colette L. Picard, Rebecca A. Povilus, Ben P. Williams, Mary Gehring
Plant stem cell organization and differentiation at single-cell resolution
James W. Satterlee, Josh Strable, Michael J. Scanlon
The lncRNA APOLO interacts with the transcription factor WRKY42 to trigger root hair cell expansion in response to cold
Michaël Moison, Javier Martínez Pacheco, Leandro Lucero, Camille Fonouni-Farde, Johan Rodríguez-Melo, Aurélie Christ, Jérémie Bazin, Moussa Benhamed, Fernando Ibañez, Martin Crespi, José M. Estevez, Federico Ariel
The conserved plant PM19 protein functions as an osmosensor and regulator of germination
Ross D. Alexander, Pablo Castillejo-Pons, Omar Alsaif, Yvonne Stahl, Madeleine Seale, Peter C. Morris
Expansin-controlled cell wall stiffness regulates root growth in Arabidopsis
Marketa Samalova, Kareem Elsayad, Alesia Melnikava, Alexis Peaucelle, Evelina Gahurova, Jaromir Gumulec, Ioannis Spyroglou, Elena V. Zemlyanskaya, Elena V. Ubogoeva, Jan Hejatko
Expression partitioning of duplicate genes at single cell resolution in Arabidopsis roots
Jeremy E. Coate, Andrew D. Farmer, John Schiefelbein, Jeff J. Doyle
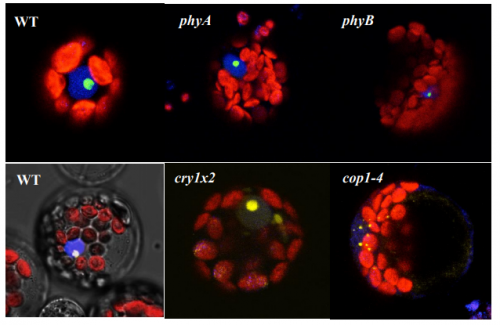
AtHDA15 attenuates COP1 via transcriptional quiescence, direct binding, and sub-compartmentalization during photomorphogenesis
Malona V. Alinsug, Custer C. Deocaris
Characterization of novel pollen-expressed transcripts reveals their potential roles in pollen heat stress response in Arabidopsis thaliana
Nicholas Rutley, Laetitia Poidevin, Tirza Doniger, Richard Tillet, Abhishek Rath, Javier Forment, Gilad Luria, Karen Schlauch, Alejandro Ferrando, Jeffery Harper, Gad Miller
Primary carbohydrate metabolism genes participate in heat stress memory at the shoot apical meristem of Arabidopsis thaliana
Justyna Jadwiga Olas, Federico Apelt, Maria Grazia Annunziata, Sarah Isabel Richard, Saurabh Gupta, Friedrich Kragler, Salma Balazadeh, Bernd Mueller-Roeber
The role of CLV1, CLV2 and HPAT homologs in nitrogen-regulation of root development
Chenglei Wang, James B Reid, Eloise Foo
The mammalian CLU homolog FMT controls development and behavior in Arabidopsis
Alexandra Ralevski, Federico Apelt, Justyna J. Olas, Bernd Mueller-Roeber, Elena I. Rugarli, Friedrich Kragler, Tamas L. Horvath
A constitutively monomeric UVR8 photoreceptor allele confers enhanced UV-B photomorphogenesis
Roman Podolec, Kelvin Lau, Timothée B. Wagnon, Michael Hothorn, Roman Ulm
Jasmonic acid coordinates with light to regulate branching angle of Arabidopsis lateral roots
Manvi Sharma, Mohan Sharma, Muhammed Jamsheer K, Ashverya Laxmi
Developmental constraints modulate reproductive fate and plasticity within the Arabidopsis ovule primordium
Elvira Hernandez-Lagana, Gabriella Mosca, Ethel Mendocilla Sato, Nuno Pires, Anja Frey, Alejandro Giraldo-Fonseca, Ueli Grossniklaus, Olivier Hamant, Christophe Godin, Arezki Boudaoud, Daniel Grimanelli, Daphné Autran, Célia Baroux
Development and cell cycle dynamics of the root apical meristem in the fern Ceratopteris richardii
Alejandro Aragón-Raygoza, Alejandra Vasco, Ikram Blilou, Luis Herrera-Estrella, Alfredo Cruz-Ramírez

SvFUL2, an A-class MADS-box transcription factor, is necessary for inflorescence determinacy in model panicoid cereal, Setaria viridis
Jiani Yang, Edoardo Bertolini, Max Braud, Jesus Preciado, Adriana Chepote, Hui Jiang, Andrea L. Eveland
Embryo CHH hypermethylation is mediated by RdDM and is autonomously directed in Brassica rapa
Tania Chakraborty, Timmy Kendall, Jeffrey W. Grover, Rebecca A. Mosher
Comparative transcriptomics identifies differences in the regulation of the floral transition between Arabidopsis and Brassica rapa cultivars
Alexander Calderwood, Jo Hepworth, Shannon Woodhouse, Lorelei Bilham, D. Marc Jones, Eleri Tudor, Mubarak Ali, Caroline Dean, Rachel Wells, Judith A. Irwin, Richard J. Morris
Spatial transcriptional signatures define margin morphogenesis along the proximal-distal and medio-lateral axes in the complex leaf of tomato (Solanum lycopersicum)
Ciera C. Martinez, Siyu Li, Margaret R. Woodhouse, Keiko Sugimoto, Neelima R. Sinha
Underground gibberellin activity: differential gibberellin response in tomato shoots and roots
Uria Ramon, David Weiss, Natanella Illouz-Eliaz
RNA-Seq analysis of genes affected by Cyclophilin A/DIAGEOTROPICA (DGT) in tomato root development
Maria G. Ivanchenko, Olivia R. Ozguc, Stephanie R. Bollmann, Valerie N. Fraser, Molly Megraw
Diverse roles of MAX1 homologues in rice
Marek Marzec, Apriadi Situmorang, Philip B. Brewer, Agnieszka Brąszewska-Zalewska
Rice embryogenic trigger BABY BOOM1 promotes somatic embryogenesis by upregulation of auxin biosynthesis genes
Imtiyaz Khanday, Christian Santos-Medellín, Venkatesan Sundaresan
Targeted knockdown of ribulose-1, 5-bisphosphate carboxylase-oxygenase in rice mesophyll cells impact on photosynthesis and growth
Chirag Maheshwari, Robert A Coe, Shanta Karki, Sarah Covshoff, Ronald Tapia, Aruna Tyagi, Julian M. Hibberd, Robert T. Furbank, W Paul Quick, Hsiang-Chun Lin
Brassinosteroids Promote Parenchyma Cell and Secondary Xylem Development in Sugar Beet (Beta vulgaris L.) Root
Wei Wang, Yaqing Sun, Guolong Li, Shaoying Zhang
ITS2 Pretrial Gene Identification Related to Seed and Flower Identification for Cyclea barbata
Monica Pignatti, William Jensen, Veronica Henderson
Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species
Jixiang Kong, Susana Martín-Ortigosa, John Finer, Nuananong Orchard, Andika Gunadi, Lou Ann Batts, Dhiraj Thakare, Bradford Rush, Oliver Schmitz, Maarten Stuiver, Paula Olhoft, David Pacheco-Villalobos
A chimera including a GROWTH-REGULATING FACTOR (GRF) and its cofactor GRF-INTERACTING FACTOR (GIF) increases transgenic plant regeneration efficiency
Juan M. Debernardi, David M. Tricoli, Maria F. Ercoli, Sadiye Hayta, Pamela Ronald, Javier F. Palatnik, Jorge Dubcovsky
Evo-devo & evo
Speciation and the developmental alarm clock
Asher D. Cutter, Joanna D. Bundus
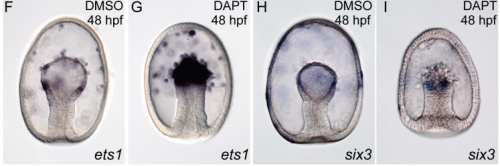
Systematic comparison of developmental GRNs explains how novelty is incorporated in early development
Gregory A. Cary, Brenna S. McCauley, Olga Zueva, Joseph Pattinato, William Longabaugh, Veronica F. Hinman
Gene expression patterns in chordate embryonic development suggest partial applicability of Haeckel’s postulates
Philipp Khaitovich, Song Guo, Haiyang Hu, Chuan Xu, Naoki Irie
Evolution of the Bicoid homeodomain required amino acid substitutions in three subdomains and epistatic interactions between them
Pinar Onal, Himari Imaya Gunasinghe, Kristaley Yui Umezawa, Michael Zheng, Jia Ling, Stephen Small
DNA methylation in cockroaches is essential in early embryo development and reduces gene expression noise
Alba Ventos-Alfonso, Guillem Ylla, Jose-Carlos Montañes, Xavier Belles
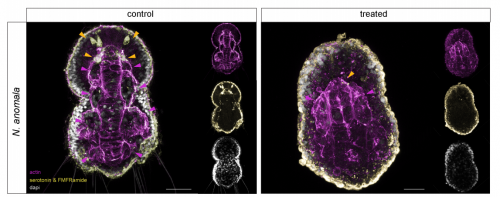
FGF signaling induces mesoderm in members of Spiralia
Carmen Andrikou, Andreas Hejnol
Molluscan dorsal-ventral patterning relying on BMP2/4 and Chordin provides insights into spiralian development and bilaterian body plan evolution
Sujian Tan, Pin Huan, Baozhong Liu
A Wnt-specific astacin proteinase controls head formation in Hydra
Berenice Ziegler, Irene Yiallouros, Benjamin Trageser, Sumit Kumar, Moritz Mercker, Svenja Kling, Maike Fath, Uwe Warnken, Martina Schnölzer, Thomas W. Holstein, Markus Hartl, Anna Marciniak-Czochra, Jörg Stetefeld, Walter Stöcker, Suat Özbek
The ontology of the anatomy and development of the solitary ascidian Ciona
Kohji Hotta, Delphine Dauga, Lucia Manni
Ciona Brachyury proximal and distal enhancers have different FGF dose-response relationships
Matthew J. Harder, Julie Hix, Wendy M. Reeves, Michael T. Veeman
Deep origins of chordate midbrain visual processing centers
Cezar Borba, Shea Schwennicke, Matthew J. Kourakis, William C. Smith
The essential role of Dnmt1 in gametogenesis in the large milkweed bug Oncopeltus fasciatus
Joshua T. Washington, Katelyn R. Cavender, Ashley U. Amukamara, Elizabeth C. McKinney, Robert J. Schmitz, Patricia J. Moore
Myomixer is expressed during embryonic and post-larval hyperplasia, muscle regeneration and fusion of myoblats in rainbow trout (Oncorhynchus mykiss)
Miquel Perrello-Amoros, Cécile Rallière, Joaquim Gutiérrez, Jean-Charles Gabillard
Cutting across structural and transcriptomic scales translates time across the lifespan and resolves frontal cortex development in human evolution
Christine J. Charvet
EMBRYONIC DEVELOPMENT OF THE FIRE-EYE-TETRA Moenkhausia oligolepis (CHARACIFORMES: CHARACIDAE)
Raquel Santos dos Santos, Jeane Rodrigues Rodrigues, Jhennifer Gomes Cordeiro, Hadda Tercya, Marissol Leite, Bruna Patrícia Dutra Costa, Raphael da Silva Costa, Caio Maximino, Diógenes Henrique de Siqueira-Silva
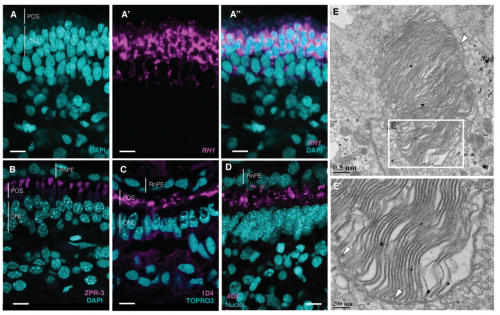
Vertebrate features revealed in the rudimentary eye of the Pacific hagfish (Eptatretus stoutii)
Emily M. Dong, W. Ted Allison
Variation in developmental rates is not linked to environmental unpredictability in annual killifishes
P. K. Rowiński, W. Sowersby, J. Näslund, S. Eckerström-Liedholm, K. Gotthard, B. Rogell
An Engrailed1 enhancer underlies human thermoregulatory evolution
Daniel Aldea, Yuji Atsuta, Blerina Kokalari, Stephen Schaffner, Bailey Warder, Yana Kamberov
Bacterial symbionts in animal development: arginine biosynthesis complementation enables larval settlement in a marine sponge
Hao Song, Olivia H Hewitt, Sandie M Degnan
Morphological and genomic shifts in mole-rat ‘queens’ increase fecundity but reduce skeletal integrity
Rachel A. Johnston, Philippe Vullioud, Jack Thorley, Henry Kirveslahti, Leyao Shen, Sayan Mukherjee, Courtney Karner, Tim Clutton-Brock, Jenny Tung
Genomics of a killifish from the Seychelles islands supports transoceanic island colonization and reveals relaxed selection of developmental genes
Rongfeng Cui, Alexandra M Tyers, Zahabiya Juzar Malubhoy, Sadie Wisotsky, Stefano Valdesalici, Elvina Henriette, Sergei L Kosakovsky Pond, Dario Riccardo Valenzano
The avian W chromosome is a refugium for endogenous retroviruses with likely effects on female-biased mutational load and genetic incompatibilities
Valentina Peona, Octavio M. Palacios-Gimenez, Julie Blommaert, Jing Liu, Tri Haryoko, Knud A. Jønsson, Martin Irestedt, Qi Zhou, Patric Jern, Alexander Suh
Sexual Reproduction in Bdelloid Rotifers
Veronika N. Laine, Timothy Sackton, Matthew Meselson
Structure and contingency determine mutational hotspots for flower color evolution
Lucas C. Wheeler, Boswell A. Wing, Stacey D. Smith
Changes in Cell Size and Shape During 50,000 Generations of Experimental Evolution with Escherichia coli
Nkrumah A. Grant, Ali Abdel Magid, Joshua Franklin, Yann Dufour, Richard E. Lenski
The order of trait emergence in the evolution of cyanobacterial multicellularity
Katrin Hammerschmidt, Giddy Landan, Fernando Domingues Kümmel Tria, Jaime Alcorta, Tal Dagan
Cell biology
Septins and a formin have distinct functions in chiral cortical rotation in the anaphase C. elegans zygote
Adhham Zaatri, Jenna A. Perry, Amy Shaub Maddox
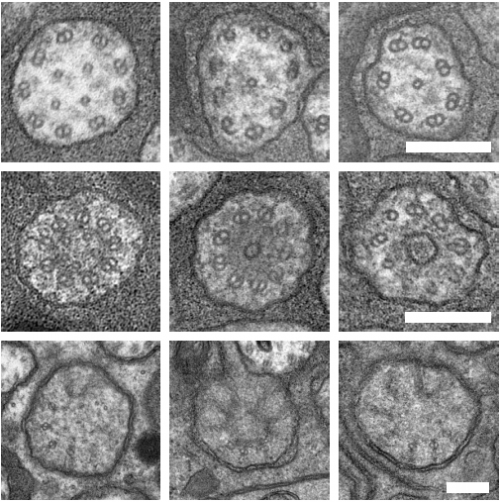
An Acentriolar Centrosome At The C. elegans Ciliary Base
Joachim Garbrecht, Triin Laos, Elisabeth Holzer, Margarita Dillinger, Alexander Dammermann
Binary decision between asymmetric and symmetric cell division is defined by the balance of PAR proteins in C. elegans embryos
Yen Wei Lim, Fu-Lai Wen, Prabhat Shankar, Tatsuo Shibata, Fumio Motegi
Centriole-less pericentriolar material serves as a microtubule organizing center at the base of C. elegans sensory cilia
Jérémy Magescas, Sani Eskinazi, Michael V. Tran, Jessica L. Feldman
Src-dependent NM2A tyrosine-phosphorylation regulates actomyosin dynamics
Cláudia Brito, Francisco S. Mesquita, Daniel S. Osório, Joana Pereira, Neil Billington, James R. Sellers, Didier Cabanes, Ana X. Carvalho, Sandra Sousa
The intrinsically disordered protein SPE-18 promotes localized assembly of the major sperm protein in C. elegans spermatocytes
Kari L. Price, Marc Presler, Christopher M. Uyehara, Diane C. Shakes
Tau, XMAP215/Msps and Eb1 jointly regulate microtubule polymerisation and bundle formation in axons
Ines Hahn, Andre Voelzmann, Jill Parkin, Judith Fuelle, Paula G Slater, Laura A Lowery, Natalia Sanchez-Soriano, Andreas Prokop
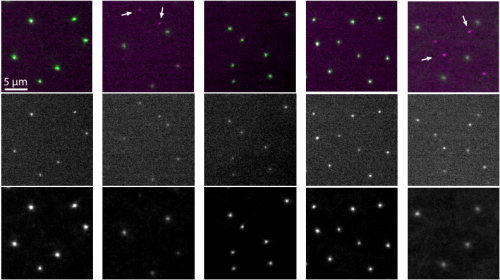
Ana1 recruits PLK1 to mother centrioles to promote mitotic PCM assembly and centriole elongation
Ines Alvarez-Rodrigo, Alan Wainman, Jordan W. Raff
Protein Phosphatase 2A-B56 maintains the meiotic spindle, kinetochore attachments and cohesion by antagonizing Aurora B in Drosophila Oocytes
Janet K. Jang, Amy C. Gladstein, Arunika Das, Zachary Sisco, Kim S. McKim
Selective dephosphorylation by PP2A-B55 directs the meiosis I – meiosis II transition in oocytes
S. Zachary Swartz, Hieu T. Nguyen, Brennan C. McEwan, Mark E. Adamo, Iain M. Cheeseman, Arminja N. Kettenbach
Celsr1 and CAMSAP3 differently regulate intercellular and intracellular cilia orientation in oviduct multiciliated cells
Fumiko Matsukawa Usami, Masaki Arata, Dongbo Shi, Sanae Oka, Yoko Higuchi, Fadel Tissir, Masatoshi Takeichi, Toshihiko Fujimori
Cell cycle-dependent active stress drives epithelia remodeling
John Devany, Daniel M. Sussman, M. Lisa Manning, Margaret L. Gardel
A unified view of neighbour cell engagement during apoptotic cell extrusion
Kinga Duszyc, Guillermo A. Gomez, Anne K. Lagendijk, Mei-Kwan Yau, Briony L. Gliddon, Thomas E. Hall, Suzie Verma, Benjamin M. Hogan, Stuart M. Pitson, David P. Fairlie, Robert G. Parton, Alpha S. Yap
Super-resolution imaging uncovers the nanoscopic segregation of polarity proteins in epithelia
Pierre Mangeol, Dominique Massey-Harroche, Fabrice Richard, Pierre-François Lenne, André Le Bivic
Involvement of uterine natural killer cells in the natural selection of human embryos at implantation
Chow-Seng Kong, Alexandra Almansa Ordoñez, Sarah Turner, Tina Tremaine, Joanne Muter, Emma S. Lucas, Emma Salisbury, Rita Vassena, Ali A. Fouladi-Nashta, Gustavo Tiscornia, Geraldine Hartshorne, Jan J. Brosens, Paul J. Brighton
Molecular Contribution to Embryonic Aneuploidy and Genotypic Complexity During Initial Cleavage Divisions of Mammalian Development
Kelsey E Brooks, Brittany L Daughtry, Brett Davis, Melissa Y Yan, Suzanne S Fei, Lucia Carbone, Shawn L Chavez
A non-canonical function for Centromere-associated protein-E (CENP-E) controls centrosome integrity and orientation of cell division
Mikito Owa, Brian Dynlacht
3D in situ imaging of female reproductive tract reveals molecular signatures of fertilizing spermatozoa in mice
Lukas Ded, Jae Yeon Hwang, Kiyoshi Miki, Huanan F. Shi, Jean-Ju Chung
Modelling
Developmental Incongruity as a Dynamical Representation of Heterochrony
Bradly Alicea
Three-Component Model of the Spinal Nerve Branching Pattern, based on the View of the Lateral Somitic Frontier and Experimental Validation
Shunsaku Homma, Takako Shimada, Ikuo Wada, Katsuji Kumaki, Noboru Sato, Hiroyuki Yaginuma
A generative network model of neurodevelopment
Danyal Akarca, Petra E Vértes, Edward T Bullmore, the CALM team, Duncan E Astle
Learning the dynamics of cell-cell interactions in confined cell migration
David B. Brückner, Nicolas Arlt, Alexandra Fink, Pierre Ronceray, Joachim O. Rädler, Chase P. Broedersz
Tools & resources
The Planarian Anatomy Ontology: A resource to connect data within and across experimental platforms
Stephanie H. Nowotarski, Erin L. Davies, Sofia M. C. Robb, Eric J. Ross, Nicolas Matentzoglu, Viraj Doddihal, Mol Mir, Melainia McClain, Alejandro Sánchez Alvarado
A Unified Framework for Lineage Tracing and Trajectory Inference
Aden Forrow, Geoffrey Schiebinger
Visualization of individual cell division history in complex tissues
Annina Denoth-Lippuner, Baptiste N. Jaeger, Tong Liang, Stefanie E. Chie, Lars N. Royall, Merit Kruse, Benjamin D. Simons, Sebastian Jessberger
An immobilization technique for long-term time-lapse imaging of explanted Drosophila tissues
Matthew P. Bostock, Anadika R. Prasad, Rita Chaouni, Alice C. Yuen, Rita Sousa-Nunes, Marc Amoyel, Vilaiwan M. Fernandes
Precise genome engineering in Drosophila using prime editing
Justin A Bosch, Gabriel Birchak, Norbert Perrimon
Melting dsDNA donor molecules potentiates precision genome editing in C. elegans
Krishna S. Ghanta, Craig C. Mello
Cas9-induced large deletions and small indels are controlled in a convergent fashion
Michael Kosicki, Felicity Allen, Allan Bradley
CRISPR Turbo Accelerated Knock Out (CRISPy TAKO) for rapid in vivo screening of gene function
Sonja L Plasil, Amit Seth, Gregg E Homanics
Knock-in of labeled proteins into 5’UTR enables highly efficient generation of stable cell lines
Faryal Ijaz, Koji Ikegami
Bioorthogonal red and far-red fluorogenic probes for wash-free live-cell and super-resolution microscopy
Philipp Werther, Klaus Yserentant, Felix Braun, Kristin Grussmayer, Vytautas Navikas, Miao Yu, Zhibin Zhang, Michael J. Ziegler, Christoph Mayer, Antoni J. Gralak, Marvin Busch, Weijie Chi, Frank Rominger, Aleksandra Radenovic, Xiaogang Liu, Edward A. Lemke, Tiago Buckup, Dirk-Peter Herten, Richard Wombacher
Highly efficient genome modification of cultured primordial germ cells with lentiviral vectors to generate transgenic songbirds
Ivana Gessara, Falk Dittrich, Moritz Hertel, Staffan Hildebrand, Alexander Pfeifer, Carolina Frankl-Vilches, Mike McGrew, Manfred Gahr
A non-invasive method to generate induced pluripotent stem cells from primate urine
Johanna Geuder, Mari Ohnuki, Lucas E. Wange, Aleksandar Janjic, Johannes W. Bagnoli, Stefan Müller, Artur Kaul, Wolfgang Enard
High-speed, long-term, 4D in vivo lifetime imaging in intact and injured zebrafish and mouse brains by instant FLIM
Yide Zhang, Ian H. Guldner, Evan L. Nichols, David Benirschke, Cody J. Smith, Siyuan Zhang, Scott S. Howard
CRISPR-Cas9 integrates exogeneous sorting new recombinant DNA in the model tree Populus trichocarpa
Ali Movahedi, Hui Wei, Zhong-Hua Chen, Weibo Sun, Jiaxin Zhang, Dawei Li, Honghua Ruan, Qiang Zhuge
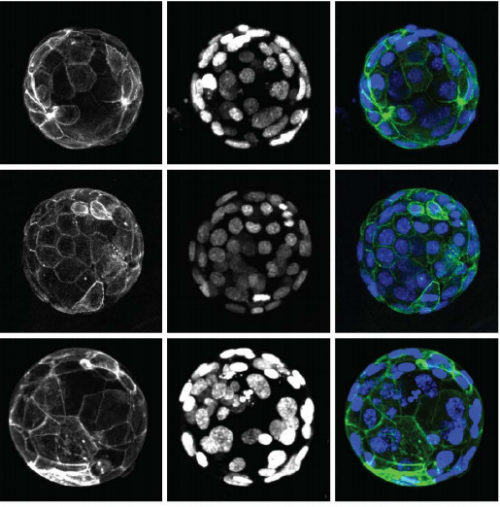
Efficient generation of endogenous protein reporters for mouse preimplantation embryos
Dan O’Hagan, Amy Ralston
CAS-LiveFISH: Simple and versatile imaging of genomic loci in live mammalian cells and early pre-implantation embryos
Yongtao Geng, Alexandros Pertsinidis
A toolkit to generate inducible and interconvertible Drosophila transgenes
Franz Wendler, Sangbin Park, Claire Hill, Alessia Galasso, Kathleen R. Chang, Iman Awan, Yulia Sudarikova, Mar Bustamante, Sichen Liu, Ethan Sung, Gabrielle Aisabonoko, Seung K. Kim, Luis Alberto Baena-Lopez
Identification of split-GAL4 drivers and enhancers that allow regional cell type manipulations of the Drosophila melanogaster intestine
Ishara S. Ariyapala, Jessica M. Holsopple, Ellen M. Popodi, Dalton G. Hartwick, Lily Kahsai, Kevin R. Cook, Nicholas S. Sokol
Development of a pan-neuronal genetic driver in Aedes aegypti mosquitoes
Zhilei Zhao, David Tian, Carolyn S. McBride
Behavioral assays to study neural development in Xenopus laevis tadpoles
Arseny S. Khakhalin, Virgilio Lopez III, Carlos Aizenman
TRAP-based allelic translation efficiency imbalance analysis to identify genetic regulation of ribosome occupancy in specific cell types in vivo.
Yating Liu, Anthony D. Fischer, Celine L. St. Pierre, Juan F. Macias-Velasco, Heather A. Lawson, Joseph D. Dougherty
Tissue-specific transcription footprinting using RNA PoI DamID (RAPID) in C. elegans
Georgina Gomez-Saldivar, Jaime Osuna-Luque, Dominique A Glauser, Sophie Jarriault, Peter Meister
An optogenetic method for interrogating YAP1 and TAZ nuclear-cytoplasmic shuttling
Anna M Dowbaj, Robert P Jenkins, John M Heddleston, Alessandro Ciccarelli, Todd Fallesen, Klaus Hahn, Marco Montagner, Erik M Sahai
Simple RGC: ImageJ plugins for counting retinal ganglion cells and determining the transduction efficiency of viral vectors in retinal wholemounts
Tiger Cross, Rasika Navarange, Joon-Ho Son, William Burr, Arjun Singh, Kelvin Zhang, Miruna Rusu, Konstantinos Gkoutzis, Andrew Osborne, Bart Nieuwenhuis
Measuring the average cell size in cellular tissues using Fourier Transform
Tess Homan, Sylvain Monnier, Cécile Jebane, Hélène Delanoe-Ayari
Multi-modal Nonlinear Optical and Thermal Imaging Platform for Label-Free Characterization of Biological Tissue
Wilson R Adams, Brian Mehl, Eric Lieser, Manqing Wang, Shane Patton, Graham A Throckmorton, J Logan Jenkins, Jeremy B Ford, Rekha Gautam, Jeff Brooker, E. Duco Jansen, Anita Mahadevan-Jansen
Research practice & education
Does retraction after misconduct have an impact on citations? A pre-post study
Cristina Candal-Pedreira, Alberto Ruano-Ravina, E Fernández, Jorge A. Ramos-Castaneda, I Campos-Varela, Mónica Pérez-Ríos
A Reaction Norm Perspective on Reproducibility
Bernhard Voelkl, Hanno Würbel
Gender (im)balance in citation practices in cognitive neuroscience
Jacqueline M. Fulvio, Ileri Akinnola, Bradley R. Postle
Specialized terminology limits the reach of new scientific knowledge
Alejandro Martínez, Stefano Mammola
Survey of Core Facilities shows the importance of communication and management for optimal research quality
Isabelle C Kos, Bjoern Gerlach, Claudia Pitzer


 (No Ratings Yet)
(No Ratings Yet) In the latest episode of Genetics Unzipped, Dr Kat Arney takes a look at the progress that’s been made in tackling rare genetic disorders (and the challenges that remain) and we hear from a prenatal genetic counsellor about how new tests are helping people carrying genetic variations make decisions about starting a family.
In the latest episode of Genetics Unzipped, Dr Kat Arney takes a look at the progress that’s been made in tackling rare genetic disorders (and the challenges that remain) and we hear from a prenatal genetic counsellor about how new tests are helping people carrying genetic variations make decisions about starting a family.
 (1 votes)
(1 votes) Our lab investigates mechanisms of chromatin regulation in early mammalian development. We also study how chromatin regulation and other mechanisms are important for transposable element (TE) expression and function.
Our lab investigates mechanisms of chromatin regulation in early mammalian development. We also study how chromatin regulation and other mechanisms are important for transposable element (TE) expression and function.