Here are the highlights from the current issue of Development:
Btbd7 branches out across multiple organs
Dynamic changes in epithelial cell-cell adhesion and motility are crucial for branching morphogenesis – the process by which highly branched epithelial organs, such as the lung and kidney, grow and develop from a simple epithelial bud. Although many key regulatory factors involved in branching morphogenesis have been identified, it remains unclear how they coordinate to control epithelial cell dynamics.
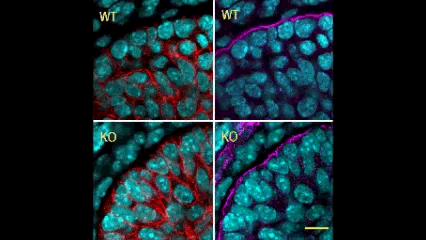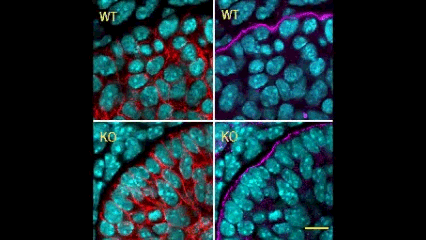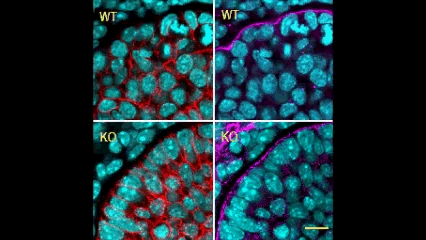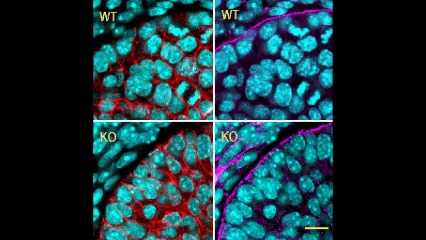
In this issue (p. 2200), the Yamada laboratory presents strong evidence for the essential role of Btbd7 in orchestrating epithelial remodelling during branching morphogenesis in the mouse lung, kidney and salivary glands in vivo. Specifically, the authors show that loss of Btbd7 in a knockout mouse model results in increased cadherin localisation to cell-cell junctions and reduced motility in the outer cells of epithelial buds. Conversely, Btbd7 overexpression in vitro leads to decreased cell-cell adhesion and increased cell motility. They further show that Btbd7 induces E-cadherin ubiquitination, internalisation and degradation in MDCK epithelial cells, providing a mechanism for how Btbd7 can control cell adhesion and migratory behaviour. Collectively, these data establish for the first time that the new regulatory molecule Btbd7 is required for successful in vivo branching morphogenesis of salivary gland, lung and kidney.
Female fertility: no bed of ROS(es)
Oocyte development involves extensive transcriptional and epigenetic changes in order to eventually produce a mature egg competent for fertilisation. Key to this process is the avoidance of apoptosis, but how anti-apoptotic genes are regulated during oogenesis remains largely undefined. Now, on p. 2165, Qinghua Shi and colleagues show that genetic deletion of histone acetyltransferase KAT8 specifically in mouse oocytes at the primordial follicle stage causes the defective development of follicles from the secondary follicle stage, which subsequently leads to female infertility. The authors observe significantly increased reactive oxygen species (ROS) levels in the mutant oocytes, coincident with the downregulation of several antioxidant genes and show via chromatin immunoprecipitation assays that KAT8 regulates antioxidant gene expression by direct binding to promoter regions. Importantly, the authors are able to rescue the defects of folliculogenesis after Kat8 deletion in oocytes by antioxidant administration in mice. This study demonstrates for the first time that KAT8 represses ROS levels in oocytes by promoting the expression of antioxidant genes at the transcriptional level, and provides insight into the epigenetic regulation of female fertility.
Now, on p. 2165, Qinghua Shi and colleagues show that genetic deletion of histone acetyltransferase KAT8 specifically in mouse oocytes at the primordial follicle stage causes the defective development of follicles from the secondary follicle stage, which subsequently leads to female infertility. The authors observe significantly increased reactive oxygen species (ROS) levels in the mutant oocytes, coincident with the downregulation of several antioxidant genes and show via chromatin immunoprecipitation assays that KAT8 regulates antioxidant gene expression by direct binding to promoter regions. Importantly, the authors are able to rescue the defects of folliculogenesis after Kat8 deletion in oocytes by antioxidant administration in mice. This study demonstrates for the first time that KAT8 represses ROS levels in oocytes by promoting the expression of antioxidant genes at the transcriptional level, and provides insight into the epigenetic regulation of female fertility.
Sox2+ pituitary stem cells on the MAP(K)
The MAPK/ERK pathway plays an important role in development and disease, with control over multiple cellular processes such as proliferation, differentiation and apoptosis, to name but a few. Despite its importance, little is known regarding its role in pituitary development and, specifically, in regulating the Sox2+ stem cell population therein.  Now, on p. 2141, Scott Haston, from the Martinez-Barbera laboratory, and colleagues take a genetic approach to address the function of the MEK/ERK pathway during normal pituitary development, and relate this to human papillary craniopharyngioma (PCP), a form of benign but clinically relevant pituitary tumour. Using a MAPK gain-of-function mouse model, the authors demonstrate that constitutive activation of the MAPK/ERK pathway during pituitary development causes pituitary hyperplasia, abnormal morphogenesis and abnormal endocrine cell specification. This is due to sustained proliferation of the Sox2+ stem cell compartment, which results in an imbalance between proliferation and differentiation and, ultimately, an expansion of Sox2+ cells at the end of gestation. Looking at human PCP samples, the authors provide evidence that sustained proliferation of SOX2+ cells with reduced differentiation potential may contribute to the underlying pathogenesis of PCP. This study is an important step forward in understanding the role of the MAPK/ERK pathway in pituitary development, and sheds light on the possible pathogenesis of human PCP.
Now, on p. 2141, Scott Haston, from the Martinez-Barbera laboratory, and colleagues take a genetic approach to address the function of the MEK/ERK pathway during normal pituitary development, and relate this to human papillary craniopharyngioma (PCP), a form of benign but clinically relevant pituitary tumour. Using a MAPK gain-of-function mouse model, the authors demonstrate that constitutive activation of the MAPK/ERK pathway during pituitary development causes pituitary hyperplasia, abnormal morphogenesis and abnormal endocrine cell specification. This is due to sustained proliferation of the Sox2+ stem cell compartment, which results in an imbalance between proliferation and differentiation and, ultimately, an expansion of Sox2+ cells at the end of gestation. Looking at human PCP samples, the authors provide evidence that sustained proliferation of SOX2+ cells with reduced differentiation potential may contribute to the underlying pathogenesis of PCP. This study is an important step forward in understanding the role of the MAPK/ERK pathway in pituitary development, and sheds light on the possible pathogenesis of human PCP.
A new model for single-cell delamination
Epithelial cell delamination – the process by which individual epithelial cells detach from an epithelial layer – is a common phenomenon throughout development and can be observed across a wide range of species. Despite its prevalence, it remains unclear whether the mechanism that drives single-cell delamination varies according to the context and, if so, how.
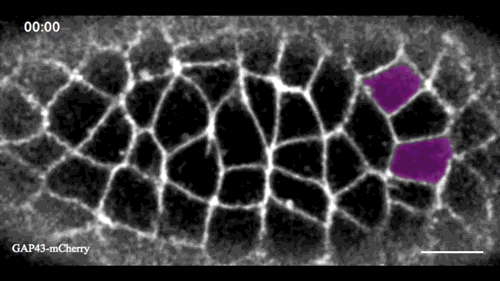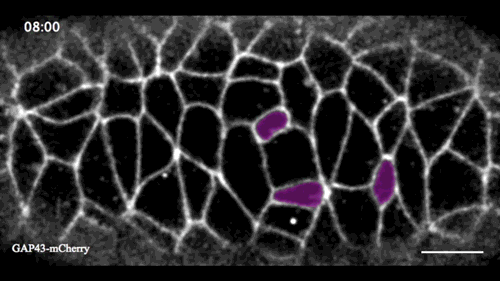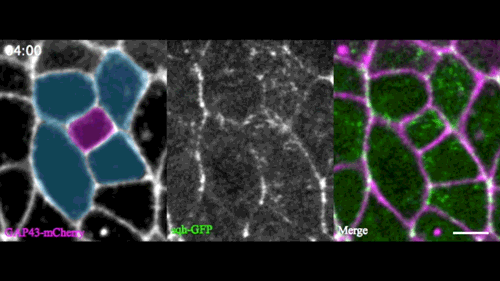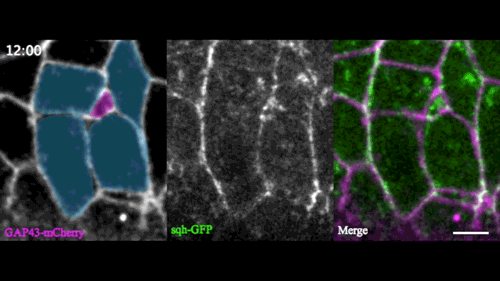
In this issue (p. 2153), Yan Yan and colleagues investigate the mechanism of neuroblast delamination in the Drosophila embryo and find that it differs to that previously reported for epithelial homeostasis. Using an elegant combination of live imaging, genetics, pharmacology and mathematical modelling, the authors describe a new mechanism for cell delamination in which the neuroblasts undergo incremental apical constriction, which coincides with pulses of myosin accumulation at their medial apical cortex. By contrast, the accumulation of junctional myosin only has a weak correlation with apical constriction.
The authors demonstrate that the quantitative difference in the frequency and magnitude of myosin pulses critically determines whether a cell will effectively constrict, and further provide evidence for the possibility that this is regulated by the same signal that defines the neuroblast cell fate, namely, Notch. This study establishes a mechanism for cell delamination that is distinct from the previously reported mechanism, and opens up a new area of research into the possibility of Notch signalling upstream of dynamic cytoskeletal rearrangements.
MSPd signalling: new roles in muscle and reproduction
Amyotrophic lateral sclerosis (ALS), which is also known as Lou Gehrig’s disease and motor neurone disease (MND), is a debilitating disease that causes the death of both cortical and spinal motor neurons, resulting in a loss of control over voluntary muscle. There is currently no cure for ALS, and the origin of the disease is largely unknown. In previous studies, VAMP/synaptobrevin-associated proteins (VAPs) have been associated with ALS and spinal muscular atrophy (SMA): specifically, the N-terminal major sperm protein domain (MSPd), which serves as an extracellular signalling molecule and which is mutated in some forms of familial ALS. In this issue, Sung Min Han, Michael Miller and colleagues investigate the molecular framework and functional consequences of MSPd signalling in C. elegans, with implications for muscle and gonad development.
In the first study (p. 2175), the authors focus on the role of MSPd in muscle formation, based on the expression of VPR-1, the C. elegans homologue of human VAPB. 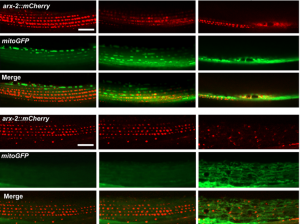 The authors show that secreted MSPds promote the localisation of mitochondria within the body wall muscle during development, and that this is dependent on signalling events that involve the CLR-1 Lar-like phosphatase receptor. Using a targeted RNAi screen of known genes implicated in ALS and SMA, the authors identified survival of motor neuron 1 (SMN-1) as a crucial downstream mediator of MSPd and further showed that SMN-1 and ARX-2 are important for mitochondria localisation along the I-bands in body wall muscle.
The authors show that secreted MSPds promote the localisation of mitochondria within the body wall muscle during development, and that this is dependent on signalling events that involve the CLR-1 Lar-like phosphatase receptor. Using a targeted RNAi screen of known genes implicated in ALS and SMA, the authors identified survival of motor neuron 1 (SMN-1) as a crucial downstream mediator of MSPd and further showed that SMN-1 and ARX-2 are important for mitochondria localisation along the I-bands in body wall muscle.
In the second study (p. 2187), the authors turn their attention to the role of VPR-1 in gonad development, an area in which the role of VPR-1 remains largely unexplored. Taking a genetic approach, the authors show that vpr-1 null mutants are sterile upon hatching, a defect in gonadogenesis that can be rescued by the expression of MSPd from almost any tissue, except for the somatic gonad itself. 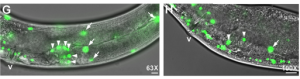 The authors further demonstrate that, under normal conditions, gonad development depends on germline and neuronal expression of vpr-1 and specifically on cleaved MSPd, which circulates in the pseudocoelom and induces gonadogenesis non-cell-autonomously, much like a hormone.
The authors further demonstrate that, under normal conditions, gonad development depends on germline and neuronal expression of vpr-1 and specifically on cleaved MSPd, which circulates in the pseudocoelom and induces gonadogenesis non-cell-autonomously, much like a hormone.
Together, these two studies represent a major step forward in our understanding of the function of the vpr-1 gene in C. elegans, and also demonstrate the utility of this organism in elucidating disease mechanisms.
Plus…
This Meeting Review of the 2017 RIKEN-CDB Symposium summarises recent progress in our understanding of human development, genetics and our evolutionary history.
This Review discusses the mechanisms that underpin the formation of skeletal muscle during development and its recapitulation from pluripotent stem cells in vitro.
This Review discusses the principal mechanisms involved in thyroid organogenesis, highlighting the factors involved in thyroid progenitor specification and the events occurring during thyroid gland morphogenesis.
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)
 (1 votes)
(1 votes)


 Now, on p.
Now, on p.  Now, on p.
Now, on p. 
 The authors show that secreted MSPds promote the localisation of mitochondria within the body wall muscle during development, and that this is dependent on signalling events that involve the CLR-1 Lar-like phosphatase receptor. Using a targeted RNAi screen of known genes implicated in ALS and SMA, the authors identified survival of motor neuron 1 (SMN-1) as a crucial downstream mediator of MSPd and further showed that SMN-1 and ARX-2 are important for mitochondria localisation along the I-bands in body wall muscle.
The authors show that secreted MSPds promote the localisation of mitochondria within the body wall muscle during development, and that this is dependent on signalling events that involve the CLR-1 Lar-like phosphatase receptor. Using a targeted RNAi screen of known genes implicated in ALS and SMA, the authors identified survival of motor neuron 1 (SMN-1) as a crucial downstream mediator of MSPd and further showed that SMN-1 and ARX-2 are important for mitochondria localisation along the I-bands in body wall muscle. The authors further demonstrate that, under normal conditions, gonad development depends on germline and neuronal expression of vpr-1 and specifically on cleaved MSPd, which circulates in the pseudocoelom and induces gonadogenesis non-cell-autonomously, much like a hormone.
The authors further demonstrate that, under normal conditions, gonad development depends on germline and neuronal expression of vpr-1 and specifically on cleaved MSPd, which circulates in the pseudocoelom and induces gonadogenesis non-cell-autonomously, much like a hormone.