Can you handle the tooth?
Posted by Kyle Martin, on 23 January 2017
Reflections on “Sox2+ progenitors in sharks link taste development with the evolution of regenerative teeth from denticles”, PNAS 113(51), 14769-14774, 2016.
Despite an overwhelming amount of carefully curated data, such as the International Shark Attack File, which indicates that your chances of being bitten by a shark are vanishingly small, humans have had a long and often macabre obsession with the impressive and sometimes daunting dentition of these stealthy marine predators. The diversity in form and function of the dentition of both living and extinct chondrichthyans (the lineage of cartilaginous fishes which includes sharks, skates, rays and chimaeras) is truly striking (some examples here). The gargantuan extinct selachian Charcharodon megalodon (literally “big tooth”) is known to science only from fossils of its immense triangular serrated teeth measuring up to 7 inches long, dwarfing the 3 inch teeth of its extant but comparatively diminutive cousin the “great” white shark C. carcharias which can nonetheless handily take down a 200kg cape fur seal (see here). In stark contrast to the sharp, often multi-cuspid, grasping and tearing tooth arrays of pelagic sharks, batoids (skates and rays) have assemblages of flat “pavement” teeth for crushing hard-shelled benthic prey (see here) while the quirky holocephalans (chimaeras/ghost sharks/rabbitfishes) have forsaken the tooth array entirely and evolved a highly derived dentition comprising relatively few large crushing tooth-plates.
Without understating the intrinsic and scientific value of this impressive morphological diversity, to an evolutionary developmental biologist the elasmobranch dentition is possessed of an even more impressive trait. This trait, if properly understood, is also one which has potentially broad developmental and biomedical applications. Unlike humans who normally produce a limited quota of exactly two generations of teeth and subsequently suffer with the consequences of tooth decay, injury, or loss, the teeth of sharks are capable of lifelong whole-organ regeneration. What’s more, sharks don’t wait for a tooth to be lost or damaged to initiate the regeneration of a replacement, but rather perpetually regenerate multiple teeth which lie in wait embedded within the jaw forming what has been endearingly referred to as the ‘conveyor-belt’ dentition. As it turns out, the capacity for endless tooth regeneration, also known as polyphyodonty, is the primitive condition of gnathostomes (jawed vertebrates) but has been lost secondarily in multiple lineages including eutherian mammals. Frustratingly for most developmental biologists, polyphyodonty has actually been lost from every major ‘classical’ model vertebrate normally studied in the lab including chick, frog, mouse and zebrafish, leaving this impressive developmental spectacle shrouded in mystery. In order to begin unravelling the secrets of polyphyodonty, we have therefore turned to the emerging model world (or simply “nature” as an evolutionary biologist would call it) and are working alongside a small but dedicated community towards turning the unassuming but satisfactorily dentate catshark, Scyliorhinus canicula, into a first-class model system. This humble but abundant shark, native to coastal waters of Europe and North Africa, was first pioneered as a model for Evo-Devo research nearly a decade ago and has since found its way into a number of Evo-Devo labs around the world including Gareth Fraser’s research group at the University of Sheffield where we maintain both developing embryos and young animals in temperature controlled artificial seawater tanks.
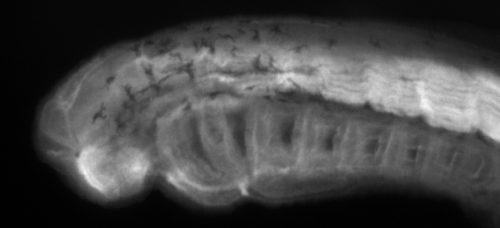
Embryos of S. canicula cultured in artificial seawater in the laboratory. After external gill eruption and the initiation of pharyngeal respiration embryos can easily be sortied from their eggcases, manipulated, and returned. Video courtesy of Dr. Andrew Gillis at the University of Cambridge.
As a postdoc in the Fraser group, I share two overarching research goals with the rest of the team. First, I believe that if we could understand the molecular developmental mechanisms regulating the perpetual regeneration of teeth in sharks and other polyphyodonts, and discover why and how this ability has been attenuated during the evolution of other lineages including our own, it may be possible to rescue this ability and stimulate tooth regeneration in humans. Second, as chondrichthyans diverged from the lineage leading to modern osteichthyans (bony fishes, including tetrapods) some 450 million years ago, they offer a deep comparative reference point as close to last common gnathostome ancestor and the ultimate evolutionary origin of teeth as one can get by comparing extant species. By comparing the expression of developmental genes in shark’s teeth with those of their bony cousins including us, we have already been able to show that many key components of the gene regulatory network used to build teeth are conserved between these groups, reinforcing the prevailing view of the biological homology of oral teeth. However, as to the ultimate evolutionary origin of the oral dentition the field has long suffered from a surfeit of theories and a deficit in data…until now.
Perhaps counterintuitively, the evolutionary history of teeth probably did not begin in the mouth. Palaeontologists and comparative anatomists have long recognized that teeth, comprised of an enamel/enameloid exterior layer, dentine core, and a bony root, are just one variety of a larger family of epithelial organs comprised of exactly the same components called odontodes. Long before there were even jaws which could conceivably house teeth, other types of odontodes made up a significant component of the defensive armour of many lineages of agnathan (jawless) vertebrates. It has therefore long been suspected that teeth originated though either an ‘expansion’ or ‘migration’ of external odontodes (known as denticles in elasmobranchs) into the mouth, after the origin of jaws. While Wolf-Ernst Reif correctly pointed out over 35 years ago that there is no histological or developmental gradation between the external denticles comprising the shark squamation and the oral dentition, this model can be resurrected in the light of modern network-based evolutionary developmental theory. In contemporary parlance, we could hypothesize the ‘co-option’ of the developmental mechanisms for producing odontodes (originally evolved in the external body armour at least according to the “outside-in” flavour of this model) into the oral cavity. According to this hypothesis it would only require the switching on of one or a few key master regulatory genes for odontode development in new competent cells of a different epithelium (of either endodermal or ectodermal origin) to initiate the entire developmental cascade for odontode production. Indeed, as we describe in our most recent paper, we found support for this model in that both denticles and teeth share equivalent expression patterns of many core developmental genes. That is, in addition to their equivalent anatomical components, they share a core conserved odontode gene regulatory network (oGRN) and should be considered as deeply homologous sensu Shubin et al. 2009. Individual teeth and denticles both develop via reciprocal signalling between an epithelial placode and underlying mesenchymal condensation, progress through comparable bud, papilla and morphogenesis stages during development, and are almost indistinguishable one from the other even at the molecular level. What then is a tooth except for an odontode in the mouth specialized for feeding?
Reconstruction of an XRM (X-Ray Microscopy) scan of a recently hatched specimen of S. canicula. The dentine/enameloid cusps of both teeth and denticles are shown in red, while the bony roots are rendered in green. The superficial skin is shown in blue and rendered semitransparent. Within the mouth the taste buds are visible as small blue mounds. Thanks to the amazing team at the London NHM Imaging and Analysis Centre for carrying out this work with us!
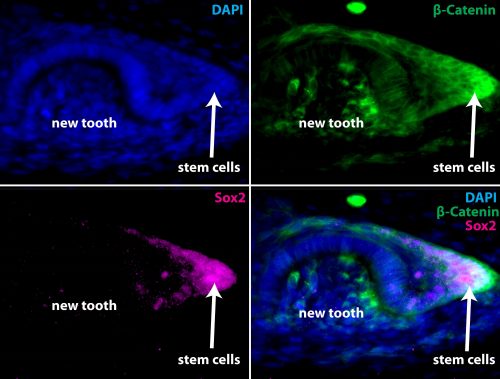
As we show in our paper, the answer to this question takes us full circle back to the most impressive property of the elasmobranch dentition: regeneration. While teeth regenerate rapidly and continuously in highly organized family rows well ahead of function, denticles do not. Using a combination of developmental gene expression analyses, cell proliferation and label-retaining analyses with the nucleoside analog BrdU, and lineage tracing with the vital dye DiI, we were able to show, at least in part, why. To make a long story (…catsharks can take upwards of 8 months to develop…) short it turns out that despite a largely conserved oGRN between teeth and denticles, the well-known stem cell factor, sox2, is only expressed in teeth. We traced the expression of sox2 from the earliest initiation of teeth through at least 5 generations and have shown that it is always retained in a small but crucial subset of epithelial cells embedded within the dental lamina immediately adjacent to the site of new tooth formation. No such pocket of set-aside stem cells could be found in denticles. Perhaps more revealing however was that the same field of sox2+ progenitors from whence the dentition emerges also gives rise to the anterior most taste-bud fields in both the upper and lower jaws. Previously, sox2 had been described as a marker of the odontogenic band, or OB, in mice and it is known to mark the future site of tooth formation in several osteichthyans along with other markers such as shh and bmp4. However, while mammals have secondarily disassociated their dentition from their gustatory faculties, many other vertebrates including sharks and teleost fishes retain a close functional and as we now know, developmental, association between teeth and taste buds. Therefore, in sharks both teeth and the anterior-most taste bud fields arise from a common sox2+ progenitor field which we have termed the odontogustatory band, or OGB, to reflect its dual developmental fate. Once this dual fate is shown definitively in an osteichthyan this will strengthen the case that this represents the ancestral gnathostome condition and reinforce the view that mammals are highly derived in this character.
In the vertebrate lineage, taste buds are comparatively ancient epithelial sensory organs, and their origin predates that of both teeth and jaws as evidenced by their presence in living petromyzontifomes (lampreys). For as long as taste buds have existed it also appears that they have been capable of regeneration, a phenomenon which unlike polyphyodonty has actually been conserved in mammals, and is known to rely deeply on the stem cell factor sox2. Taken together these data point towards an intriguing and unexpected evolutionary scenario, namely that teeth evolved through a co-option of the oGRN from non-oral odontodes within the sox2+ progenitors of the oral cavity previously directed towards forming an entirely different type of epithelial organ – taste buds. This may not have been as grand a leap for these cells to take as it might appear. In fact, many epithelial organs utilize many of the same developmental genetic blueprints to coordinate their development. There would be far fewer evolutionary steps to take in order to modify a pre-existing morphogenetic field competent to produce one type of epithelial organ (taste buds) with the plans for a different type (odontodes), than having to start from scratch in a cell type which wouldn’t normally give rise to these types of organs. Evolution is lazy, after all.
If this explanation seems somehow incomplete to you don’t worry, that only indicates that you were paying attention! Certainly we have only scratched the surface of this complex problem and there are many more questions to answer. What is this hypothetical master regulatory gene which can switch on the whole odontode differentiation pathway in a new competent epithelium? How do these common sox2+ progenitors know when to make teeth vs. taste buds and how many of each? Watch this space for more exciting developments as new data pours in from sharks and other emerging models in key phylogenetic positions.
Kyle Martin



 (7 votes)
(7 votes) from a recipient of a Company of Biologists Travelling Fellowship.
from a recipient of a Company of Biologists Travelling Fellowship. I am originally a Finnish evolutionary biology student who got fascinated about developmental biology during my exchange programme couple of years ago in Southern Chile (
I am originally a Finnish evolutionary biology student who got fascinated about developmental biology during my exchange programme couple of years ago in Southern Chile (











 (7 votes)
(7 votes)


 Naïve pluripotent mouse embryonic stem cells (ESCs) resemble the preimplantation epiblast and efficiently contribute to chimaeras. By contrast, primate ESCs correspond to the postimplantation embryo and fail to resume development in chimaeric assays. In their
Naïve pluripotent mouse embryonic stem cells (ESCs) resemble the preimplantation epiblast and efficiently contribute to chimaeras. By contrast, primate ESCs correspond to the postimplantation embryo and fail to resume development in chimaeric assays. In their  The planar cell polarity (PCP) pathway is best known for its role in polarizing epithelial cells within the plane of a tissue but it also plays a role in a range of cell migration events during development. In their
The planar cell polarity (PCP) pathway is best known for its role in polarizing epithelial cells within the plane of a tissue but it also plays a role in a range of cell migration events during development. In their  (No Ratings Yet)
(No Ratings Yet)