The people behind the papers: Joseph Pickering & Matthew Towers
Posted by the Node Interviews, on 25 October 2016
So far in this series, we’ve featured fly nuclear pores, lizard tails, squid eyes and mouse digits, and heard from researchers working in Germany, the US and Canada. Today, we switch model system and geographical location once again. The work was published recently in Development, and uses timed inhibition of sonic hedgehog signalling during chick wing development to test models of digit patterning. The people behind the paper are its two authors: postdoc Joseph Pickering and PI Matthew Towers, MRC Career Development Fellow at The University of Sheffield in the UK.

So Matt, can you tell us how you came to form your lab, and what questions your team is aiming to answer?
MT From my days as a PhD student working on the functions of cell cycle genes during snapdragon leaf development, I have been interested in how growth and pattern specification are integrated. As a postdoctoral researcher I was fortunate enough to continue working on this problem – but on chick limb development – under the supervision of Cheryll Tickle, who has contributed greatly to this field. I set up my own lab studying limb development at Sheffield, first with the support of the university and my sponsor Marysia Placzek, and then with an MRC Career Development Fellowship. I recently obtained a Wellcome Trust Senior Research Fellowship.
“The key question my lab is addressing is how cells integrate their intrinsic developmental programmes with extrinsic signals during limb patterning.”
The key question my lab is addressing is how cells integrate their intrinsic developmental programmes with extrinsic signals during limb patterning. This work has shed light on some of the classic, but controversial models of limb development – in particular, how pattern is specified along the antero-posterior (thumb-little finger) axis, and, in collaboration with Marian Ros’ lab in Santander, the proximo-distal (shoulder-digit) axis. I also co-supervise two PhD students with Marysia who are studying how growth and pattern specification are integrated during the early development of the chick hypothalamus.
Sheffield seems like an exciting place for life sciences and developmental biology at the moment?
MT The Biomedical Science department is an exciting place to study the life sciences. We also have strong links with a number of excellent research centres that focus on a range of subjects including stem cell biology, sensory neuroscience and membrane biology. I am part of a very dynamic group of researchers in the Bateson Centre who use a variety of model organisms, including zebrafish, chickens, fruit flies, slime moulds and mice to investigate a range of questions in developmental biology. These include trying to understand the basic patterning process, to elucidating how pathogens subvert the normal development of an individual.
And Joseph, how did you come to join Matthew’s lab?
JP I completed my PhD with Anne-Gaelle Borycki in Sheffield studying the regulation of an extracellular matrix protein, Laminin α1, by Sonic hedgehog (Shh) signalling in zebrafish. In fact, my paper on this has just been published.
Matt started at the university just as I was writing up my thesis. I found his work very interesting, but he was not yet ready to hire a postdoc! I left the university for a role as a scientific sales rep, which gave me a good grasp of the kinds of developmental biology research going on around the country. I kept up to date with his research activities through discussions in the pub, and in 2014, I decided to take up a postdoc position in Matt’s lab. I had developed an interest in Shh signalling and patterning mechanisms since my days as an undergraduate student, and the chick limb seemed like the ideal model to study this further.
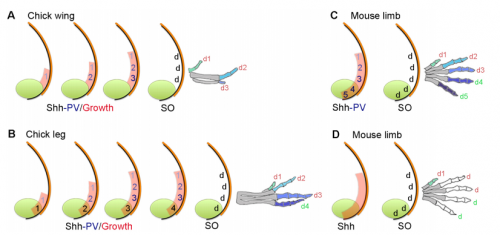
Your paper uses chicken wing development to test models of vertebrate digit patterning. What was the state of play before you started this work, particularly with regard to ideas of positional information and self-organisation?
MT&JP Positional information can explain how different digit identities (thumb vs. little finger) are specified in the chick wing. However, it has been apparent for a long time that digit number and spacing is determined by a self-organising mechanism that can be modelled by a Turing-like process. Before we started this work it was unclear how these two processes could act together to generate some of the diverse digit patterns found in nature. Thus positional information and self-organisation were often viewed as competing models of digit patterning.
“Positional information and self-organisation were often viewed as competing models of digit patterning.”
Could you talk us through the basic experimental rationale behind the paper?
JP: I was investigating another problem using the drug cyclopamine to inhibit the Shh pathway in the chick wing bud. Unexpectedly, I noticed that at a specific stage of development, the cyclopamine-treated wings produced an extra digit. Matt had occasionally seen this phenotype when he was a Postdoc in Cheryll Tickle’s lab (MT – it gave me a headache because it did not seem to make any sense) and we talked about how similar it looked to patterns of digits produced by self-organisation in the absence of positional information. The study of self-organisation in the limb has had a renewed interest, especially in recent papers from James Sharpe and Marian Ros. Therefore, since it looked like we had transformed a chick wing pattern requiring positional information, to one not requiring it, I decided to study it further. I was able to routinely obtain the phenotype and this allowed us to characterise how the temporal interplay between positional information and self-organisation can produce diverse digit patterns.
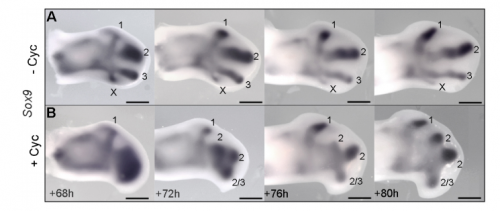
Could you sum up the key results of your paper in a paragraph?
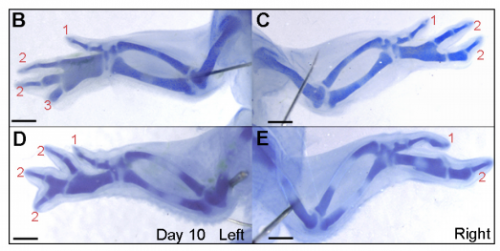
MT&JP Previously, as a postdoc I had shown that Shh signalling integrates antero-posterior growth with the specification of cells with three antero-posterior positional values that give rise to the three chick wing digits (1-2-3). In our current Development paper we showed that Shh inhibition during a very precise point during this process could unexpectedly result in wings forming with four digits (1-2-2-2, based on the pattern of phalanges). We showed that this occurred because the specification of antero-posterior positional information is truncated. However, a switch to antero-posterior growth mediated by the overlying epithelium then occurs. This expansion allows for cells specified with a digit 2 positional value to give rise to up to three digit 2s by self-organisation. Interestingly, one of these digits 2s unexpectedly arises from the Shh-producing cells of the polarizing region – an ability lost in the dinosaurian ancestors of birds.
And to expand on this theme, what does this work suggest about the evolutionary history of digit patterning?
MT&JP Previously, we had presented a model for how positional information specifies the four different positional values of the chick leg digits (1-2-3-4). As the chick leg has remained unchanged during the evolution of amniotes in terms of phalangeal pattern, we suggest that it represents the patterning mechanism for four of the five digits of the common ancestor of birds and mammals. We speculate that the ancestral mechanism has been maintained in the bird wing, apart from the loss of the most-posterior digits that arise from the polarizing region (digits 4 and 5). However, in light of our new findings, we propose that the specification of positional information has been curtailed in the mammalian limb, but that, based on phalangeal pattern, self-organisation then results in the formation of several digit 2s (1-2-2-2-2).
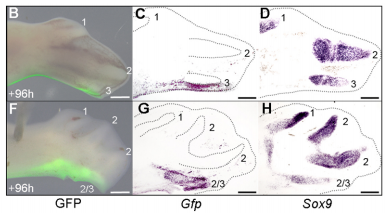
When doing the research, was there a particularly exciting result or eureka moment that stayed with you?
JP Successfully grafting a polarising region from a GFP-expressing chicken into a wild-type chicken felt like quite an achievement by itself, but did not come close to the excitement of discovering that the graft contributes to the fourth digit of cyclopamine-treated wings! This is the first piece of work to clearly show that a digit can arise from the polarizing region of the chicken wing bud, and gave important insights into how mammalian digits may be patterned. We were also surprised because we had expected an additional central digit to arise in the cyclopamine-treated wings, not an extra posterior digit. I also once managed to juggle 3 eggs for at least 10 seconds.
And what about the flipside: any particular moments or frustration and despair?
JP Performing the experimental work in this paper was very repetitive and laborious due to the sheer number of embryos that would die. This was particularly frustrating when it involved difficult experiments such as tissue grafting.
So what next for you, Joseph?
JP More chicks and drugs and rock ‘n’ roll! I’m currently trying to understand how the four-digit phenotype that I have characterised occurs at the molecular level. I am testing various candidate genes for their role in digit formation from the polarising region using chicken and mouse embryos, and also validating an RNA-sequencing screen that I have performed on cells of the developing chick limb.
More chicks and drugs and rock ‘n’ roll!
And what is the Towers lab currently working on?
MT Our recent research has highlighted the importance of cells measuring time to intrinsically execute their developmental programmes (Chinnaiya et al, Nat Comm 2014, Saiz-Lopez et al 2015). Now we are particularly interested in the molecular nature of intrinsic timing mechanisms, and how they could be used to scale pattern formation both within, and between different species.
Browse the People Behind the Papers archive here.


 (7 votes)
(7 votes) (No Ratings Yet)
(No Ratings Yet)
 Finally, the Autum Meeting hosted the inaugural Dennis Summerbell Lecture which was given by
Finally, the Autum Meeting hosted the inaugural Dennis Summerbell Lecture which was given by  Dementia causes enormous personal hardship and costs the UK ~£23 billion every year. The second most common form is Frontotemporal lobar degeneration (FTLD). About 40% of FTLD cases have genetic causes, with >8% involving abnormal aggregate-forming GA, GR, PR, GP and AP dipeptide repeat proteins (DPRs).
Dementia causes enormous personal hardship and costs the UK ~£23 billion every year. The second most common form is Frontotemporal lobar degeneration (FTLD). About 40% of FTLD cases have genetic causes, with >8% involving abnormal aggregate-forming GA, GR, PR, GP and AP dipeptide repeat proteins (DPRs).
 ch of a new Special Collection named Spotlight on Rat: Translational Impact. The rat is a key model for basic and preclinical studies of physiology, pharmacology, toxicology and neuroscience, underlining its importance in studies of human disease. There are many reasons for its suitability as a model system – the close evolutionary and genomic relationship to humans, the sophistication and sociability of the animal, the ease of physiological and behavioural measurements, and the recent proliferation of transgenic and knockout rats, enabled by new and improved technologies for genetic manipulation.
ch of a new Special Collection named Spotlight on Rat: Translational Impact. The rat is a key model for basic and preclinical studies of physiology, pharmacology, toxicology and neuroscience, underlining its importance in studies of human disease. There are many reasons for its suitability as a model system – the close evolutionary and genomic relationship to humans, the sophistication and sociability of the animal, the ease of physiological and behavioural measurements, and the recent proliferation of transgenic and knockout rats, enabled by new and improved technologies for genetic manipulation.



 The intestinal epithelium is the fastest renewing tissue in mammals and has a large flexibility to adapt to different types of damage. Here,
The intestinal epithelium is the fastest renewing tissue in mammals and has a large flexibility to adapt to different types of damage. Here, 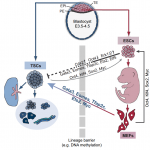 Trophoblast stem cells (TSCs) retain the capacity to self-renew indefinitely and harbour the potential to differentiate into all trophoblast subtypes of the placenta. Recent studies have shown how signalling cascades integrate with transcription factor circuits to govern the fine balance between TSC self-renewal and differentiation. In addition, breakthroughs in reprogramming strategies have enabled the generation of TSCs from fibroblasts. Here,
Trophoblast stem cells (TSCs) retain the capacity to self-renew indefinitely and harbour the potential to differentiate into all trophoblast subtypes of the placenta. Recent studies have shown how signalling cascades integrate with transcription factor circuits to govern the fine balance between TSC self-renewal and differentiation. In addition, breakthroughs in reprogramming strategies have enabled the generation of TSCs from fibroblasts. Here, 