I woke up this morning to a Facebook reminder of where I was 5 years ago. I was in Lille, France, on a 2 month sabbatical at Université Lille 1 from my PhD at the University of Cambridge, UK. It was supported by an EU collaborative grant to promote scientific interaction between member states.
By all measures, it was a roaring success – far better than we had dared to hope. I was working with one of the world’s least purifiable proteins, a transcription factor called Neurogenin2, and trying to study its phosphorylation status by NMR (nuclear magnetic resonance) imaging. We had gone in with the expectation that nothing would work but that it was worth a go, and I came out with an entire thesis chapter based on 2 months work, and what we would later publish as, “Phosphorylation in intrinsically disordered regions regulates the activity of Neurogenin2.”
Not only was it directly useful as a scientific endeavor, it was also a great experience. I learned about some of the intricacies of the French system, working with an independent researcher at CNRS who did not run the lab, but was nonetheless a researcher in her own right. My Parisian-French-with-a-Northern-Irish-accent proved useless in the face of a local dialect, Ch’ti, on the French-Belgian border. I went to Waterloo near its anniversary; I ate vast amounts of cheese; I marvelled at the holidays the French seemed to have every other week. I lived in dank student accommodation (about which my hosts were very apologetic) but I have nothing but fond memories for the 2 months I spent there. In the space of the last week, I have realised how much I took for granted that this was an opportunity available to me.
On June 23rd, the UK voted in a referendum to leave the EU.
I’m not going to comment on whether that is or isn’t going to happen, what it does or doesn’t mean for democracy in the UK – there’s enough on that already. What I’d like to do here is briefly address some areas crucial to science: funding, international collaboration and the mobility of scientific researchers. The implications for science from Brexit are both uncertain and unlikely to be clarified soon. And scientific research, as an enterprise, does not deal well with uncertainty.
Funding
Immediately after the results were certain, which I watched live, I gave my initial response and thoughts to a call from Nature for reactions from scientists:

What did I mean by “a science funding system already strained”? Last August, I made a brief visit to the UK, gave some talks on my research and scoped out the situation for academic jobs, as this was still the career track I was primarily pursuing. It became clear pretty quickly that funding was a key concern – and this was before Brexit was even really in anyone’s mind as an issue. Science funding by the UK has stagnated for years, and it was only through the major “Science is Vital” campaign that funding for science was not cut by the coalition government of 2010 – but neither was it increased. EU funding currently makes up around 15% of the UK’s research and development funding. The UK is only just behind Germany in the amount of money received from the EU for science funding and despite being a net contributor overall to the EU, has benefitted financially in terms of research funding (see Table 1 and surrounding discussion, “Examining Implications of Brexit for the UK Research Base”).
The UK has become overly dependent on EU funding – and a situation has arisen where many have allowed the EU to make up for the UK’s own lack of investment. In 2012, the UK only spent 1.63% of its GDP on research, lagging behind other countries with large research output.
The BBC reported claims from Jo Johnson, UK Science Minister, that, “world class research would “endure” in the UK following a Brexit. He added that researchers should be “optimistic about the future.” But in the same meeting he could not provide any guarantees about EU funds. The UK will also no longer be able to shape research policy from outside the EU.
The official line at the moment is that nothing has changed; the UK is still an EU member for at least two years, and there seems to be a lot of certainty that the UK will continue to pay into the Horizon 2020 and EU research funding based on its GDP, much like Switzerland. However, I have not yet seen how this is easily reconciled with a major feature of the Leave campaign – immigration – and the Swiss have restricted their access to EU funds by restricting freedom of movement, particularly funds allowing young researchers to move around, such as Marie Skłodowska-Curie Research Fellowships and the Erasmus programme (if you click this link, note the first lines on the web page).
And this is crucial because the scientific enterprise is about so much more than just money; it is about people. More concerning, in my mind, than the issue of what will happen to funding, is what the introduction of added bureaucracy and complications will do to international collaborations, and what the question over mobility of researchers – and the recent spike in xenophobia and racism – will change in the minds of junior scientists looking to come to, return to, or stay in the UK.
Collaborations
Jo Johnson, fresh from the assurances given above about UK science, soon had to address the issue that UK researchers risk exclusion from collaborative projects in the EU. This is what I cannot reconcile in my mind about the assurances that legally, nothing has changed, and everything will carry on as before. Science funding is highly competitive. To obtain that funding, researchers will seek to minimize any risk of rejection of grant applications. The UK has stated that it is not going to trigger Article 50, the mechanism to leave the EU, until the new Prime Minister is decided, at the earliest in September. EU officials have in turn retorted that no assurances or negotiations will be carried out in that time; and negotiations will then take up to 2 years.
In this vacuum devoid of certainty, with no realistic assurances, I would be very surprised if researchers in the EU, albeit with a heavy heart, do not begin to rule out collaborations with UK researchers. It is even the case that although the official line has not changed, misinformation and requests to withdraw UK researchers from or to not involve UK researchers in projects may be filtering into the system. This is already happening anecdotally:
Another area where quantitative data is sparse, but anecdotal data is also emerging, is in the mobility of young researchers.
The mobility of junior researchers
Stephen Hawking noted that the strong record the UK has in attracting EU funds, and its ability to attract young international researchers with EU grants, encouraged the best young EU scientists to think about moving to the UK. Paul Nurse has urged that free movement of people is essential for UK science, and Venkatraman Ramakrishnan, President of the Royal Society, has called for assurances for residency for EU researchers from the government.
Buzzfeed interviewed a range of scientists at various stages on their opinions and in particular these young European scientists currently in the UK who are now reconsidering their future there. There is now the question of belonging, or being welcomed, in the UK.
I titled this blog post using a direct quote from UK Justice Secretary Michael Gove, “people in this country have had enough of experts” (Michael Gove is one of the candidates for the leadership of the Conservative party, and therefore the position of Prime Minister). Xenophobia and racism have spiked recently in the UK. Reporting of hate crimes has increased. Issues of funding and academic job security aside, the UK is not presenting an attractive image to those of us outside it. There does not appear to be a desire to welcome those who are foreign or those with expertise, even in the very highest echelons of government. It is also not encouraging expats I have spoken with to return.
Anecdotal evidence exists that fellowship applications from the UK are being hit:
https://twitter.com/davecl42/status/749157977643425792
This also brings in the question of the effect on students and higher education. Current EU students and those entering in 2016 will have loan arrangements honoured, but there is no guarantee as yet with respect to entry from autumn 2017. Institutions are issuing statements along the lines of this from Vice-Chancellor Sir Leszek Borysiewicz at the University of Cambridge, and the President at University College London reacted to claims that UK universities will not suffer with fears that EU students may be lost from the UK, put off by tuition fees (students from the EU pay the same fees as UK students, and not international fees). UCL and other institutions in the UK have expressed dismay at the effects leaving the EU may have on their operation.
Concluding thoughts
In the process of negotiation, UK science and universities may end up with the relatively unchanged situation that many hope for. But that situation is a long way away, and fraught with uncertainty. Those who argue that all is fine and will continue to remain so, and that there is therefore not a problem, are not appreciating the incentives and risks that people consider as they move around to do science. Science is a highly mobile affair – look at Emmanuelle Charpentier’s movements around the world as a case in point – and the entire incentive structure for moving to the UK, and the risks involved for young researchers, have completely changed, if only temporarily, in the absence of any certainty of what is to come. Science is so competitive right now that many researchers just can’t afford to enter into uncertain positions. The longer the uncertainty continues, the more likely (and my own anecdotal evidence, admittedly small, overwhelming shows this) young scientists in the UK are to leave; and expats and foreign researchers are to stay away.
But perhaps these may not be factors that concern the UK in the end. After all, as we were told in no uncertain terms, “people in this country have had enough of experts.”
Next steps
The situation in the UK is unpredictable, to say the very least. Continuously monitoring the situation is necessary – here’s a summary of the first week, from Nature and coverage of the situation in The Atlantic for further reading.
Scientists for EU (@Scientists4EU), a campaign that previously discussed the possibilities of the effect of Brexit on science prior to the vote, are already trying to gather evidence for the effect of Brexit on UK researchers – if you have anything to contribute, know someone who does or just want to aid in data collection on this issue, please share their link on Monitoring Brexit’s impact: http://scientistsforeu.uk/monitoring-brexit-impact/
I am working on a follow-up post to this for the American Society of Cell Biology’s COMPASS blog in which I would like to include some more data as it appears, so please do help with data collection efforts.
On Tuesday also the Commons Select Committee on Science and Technology will hold a session on “Implications and opportunities for science and research examined”. You can watch at the link below:
http://parliamentlive.tv/event/index/f225ac03-d091-4215-b547-7b0050d33cca
The Node’s Question of the Month is about your opinions after the referendum – make sure to contribute there too.
Do you have thoughts or comments? Please feel free to get in touch below. Fact-checking and requests for clarification are particularly encouraged.
———————————————————————————————
Gary McDowell is the Executive of Director of Future of Research (FoR), a U.S. nonprofit that assists junior scientists in grassroots advocacy to promote solutions to problems they perceive with science, and the scientific enterprise. He spent 9 years discovering the joy of developmental biology working with the frog Xenopus laevis. He is currently a resident at the Moore Foundation-funded Manylabs open science skunkworks in San Francisco, CA.
 (7 votes)
(7 votes)
 Loading...
Loading...


 (6 votes)
(6 votes)


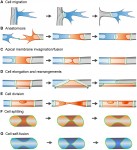 Vascular networks are formed and maintained through a multitude of angiogenic processes, such as sprouting, anastomosis and pruning. Only recently has it become possible to study the behavior of the endothelial cells that contribute to these networks at a single-cell level in vivo. Here, Markus Affolter and colleagues summarizes what is known about endothelial cell behavior during developmental angiogenesis, focusing on the morphogenetic changes that these cells undergo. See the Review on. p.
Vascular networks are formed and maintained through a multitude of angiogenic processes, such as sprouting, anastomosis and pruning. Only recently has it become possible to study the behavior of the endothelial cells that contribute to these networks at a single-cell level in vivo. Here, Markus Affolter and colleagues summarizes what is known about endothelial cell behavior during developmental angiogenesis, focusing on the morphogenetic changes that these cells undergo. See the Review on. p.  The vertebrate small intestine requires an enormous surface area to effectively absorb nutrients from food. Morphological adaptations required to establish this extensive surface include generation of an extremely long tube and convolution of the absorptive surface of the tube into villi and microvilli. In their Review,
The vertebrate small intestine requires an enormous surface area to effectively absorb nutrients from food. Morphological adaptations required to establish this extensive surface include generation of an extremely long tube and convolution of the absorptive surface of the tube into villi and microvilli. In their Review, 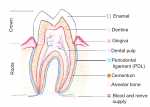 Mammalian teeth harbour mesenchymal stem cells (MSCs), which contribute to tooth growth and repair. These dental MSCs possess many in vitro features of bone marrow-derived MSCs, including clonogenicity, expression of certain markers, and following stimulation, differentiation into cells that have the characteristics of osteoblasts, chondrocytes and adipocytes. Here, Paul Sharpe outlines some recent discoveries in dental MSC function and behaviour and discusses how these and other advances are paving the way for the development of new biologically based dental therapies. See the Review on p.
Mammalian teeth harbour mesenchymal stem cells (MSCs), which contribute to tooth growth and repair. These dental MSCs possess many in vitro features of bone marrow-derived MSCs, including clonogenicity, expression of certain markers, and following stimulation, differentiation into cells that have the characteristics of osteoblasts, chondrocytes and adipocytes. Here, Paul Sharpe outlines some recent discoveries in dental MSC function and behaviour and discusses how these and other advances are paving the way for the development of new biologically based dental therapies. See the Review on p.  (No Ratings Yet)
(No Ratings Yet)

 Julia Turan wrote about the
Julia Turan wrote about the  We heard about the Young Embryologist Network meeting in London from two perspectives:
We heard about the Young Embryologist Network meeting in London from two perspectives: 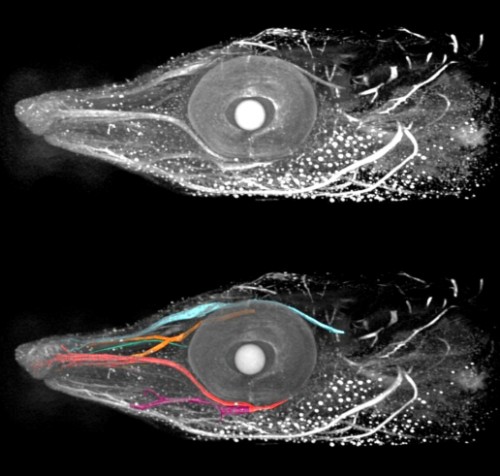 Our ‘A day in the life…’ series continued, and
Our ‘A day in the life…’ series continued, and  e asked our Questions of the Month, which really couldn’t have been about anything else: what does the UK referendum result mean to you as a scientist, and what can we as a community do about it?
e asked our Questions of the Month, which really couldn’t have been about anything else: what does the UK referendum result mean to you as a scientist, and what can we as a community do about it? 

 (7 votes)
(7 votes)
 Notch signalling regulates various aspects of vertebrate cartilage development, and Hilton and colleagues now
Notch signalling regulates various aspects of vertebrate cartilage development, and Hilton and colleagues now 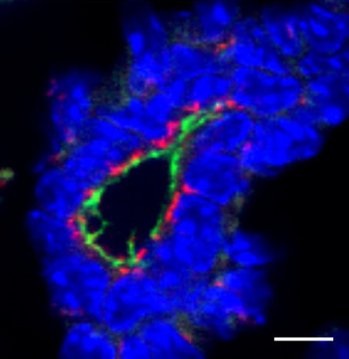 protein Scribble is critical in establishing apical-basal polarity during epithelial development. Muthuswamy and coworkers now
protein Scribble is critical in establishing apical-basal polarity during epithelial development. Muthuswamy and coworkers now 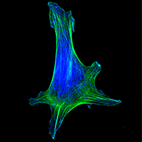 Haptotaxis is directional cell migration in response to a gradient of substrate-bound cues. Bear and colleagues
Haptotaxis is directional cell migration in response to a gradient of substrate-bound cues. Bear and colleagues 
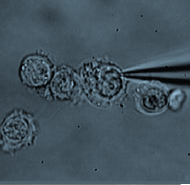 De novo generation of photoreceptor cells is therapeutically promising for patients with retinal degenerative diseases. Seko and colleagues
De novo generation of photoreceptor cells is therapeutically promising for patients with retinal degenerative diseases. Seko and colleagues 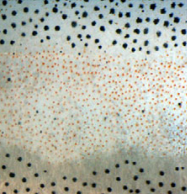 pigmentation is an established model system for developmental patterning. Irion and colleagues
pigmentation is an established model system for developmental patterning. Irion and colleagues 
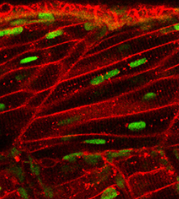 identify
identify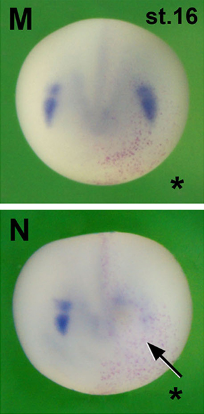 Pera and co-workers
Pera and co-workers