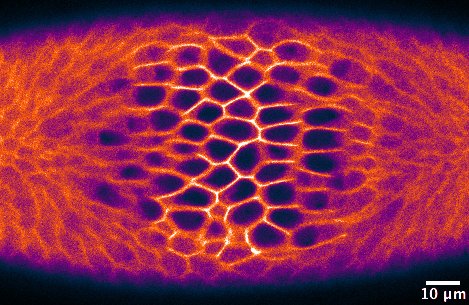
Morphogenesis, the shaping of tissues and organs, is driven by a series of events that proceed in a coordinated manner, both spatially and temporally. Such events include changes in cell shape, cell adhesion, and cell migration, which happen at a precise developmental stage in single cells or cell collectives. The detailed study of morphogenetic processes relies upon the ability to perturb these localized changes in the otherwise intact organism, ideally with sub-cellular precision and on a time-scale of seconds. However, this is impossible to achieve with traditional genetic or chemical tools.
When, in 2011, I started my PhD in the De Renzis group at EMBL, optogenetics had just been elected “Method of the Year” by Nature Methods1 and was included in Science’s “Breakthroughs of the Decade”2. Since its early approaches at the beginning of 2000s3-5, optogenetics has greatly facilitated the study of neuronal circuits in the brain. In these pioneering experiments, mice were made to express light-sensitive ion channels in selected populations of neurons, thus allowing control over brain activity with a pulse of laser light. By the end of 2000s, a few studies reported using optogenetics also in non-excitable cells6-8. Despite these advances, being able to control morphogenetic movements with light still represented a challenge.
My longstanding interest in biotechnology led me to propose co-opting optogenetics to modulate cell behaviour during Drosophila morphogenesis. Data from our lab had shown that a particular plasma membrane lipid, phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2), is essential for cell morphogenesis in the Drosophila embryo9. Indeed, PI(4,5)P2 is known to act as a molecular scaffold for actin-binding proteins, thereby regulating actin polymerization at the cell cortex. PI(4,5)P2 depletion from the plasma membrane is mediated by a class of enzymes known as inositol-polyphosphate-5-phosphatases (5-phosphatases). I reasoned that by modulating PI(4,5)P2 metabolism at the plasma membrane, I should be able to deplete actin from the cell cortex. Because cortical actin is key for many biological processes ranging from cell contractility to cell division, this tool would allow the control of a wide spectrum of morphogenetic events. The not-so-trivial question was how to do that using light.
Just a couple of years before I started my PhD, a few research groups had developed optogenetic approaches to specifically control protein-protein interaction in cell culture10-12. After considering the functional features of these tools, I decided to use the Cry2-CIB1 system12, as it can be rapidly activated with blue light and does not need the addition of an exogenous chromophore. The Cry2-CIB1 system is composed of two photosensitive modules: Cryptochrome 2 (Cry2) and CIB1, which interact only in the presence of blue light (458-488 nm). This system had already proved suitable to control phosphoinositide metabolism in cultured cells13. In the optogenetic approach I set up, a GFP-tagged CIB1 localizes to the cells’ plasma membrane, whereas the 5-phosphatase, which is fused to Cry2 and mCherry, stays in the cytosol. The fluorescent tags would allow me to monitor the protein location in vivo. To test whether I could use this system to modulate morphogenetic movements, I decided to focus on ventral furrow formation, the invagination of the Drosophila mesodermal tissue, as this process is highly dependent on actin dynamics and cell contractility. During ventral furrow formation, a stripe of ~1000 ventral cells constrict their apical surfaces and fold inwards as a tube. The core idea is that in the dark, the 5-phosphatase would remain cytosolic, and morphogenesis would proceed normally (Figure 1A). In the presence of blue light, the 5-phosphatase would relocate to the plasma membrane, where it would deplete PI(4,5)P2 and, in turn, actin from the cell cortex. This should result in the inhibition of cell contractility and a blockade of ventral furrow formation (Figure 1B).
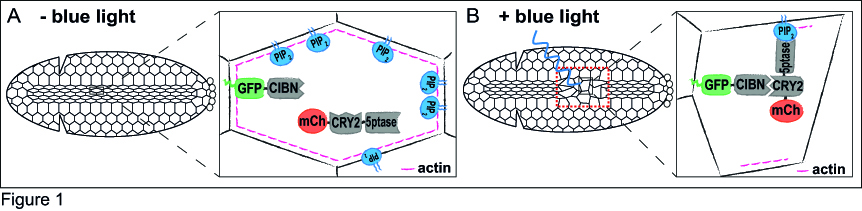
When I started to test this optogenetic approach, I first looked at the effects of blue light illumination on the 5-phosphatase localization, and on PI(4,5)P2 and actin levels. Upon a 1 s pulse of 488 nm light, the 5-phosphatase relocated to the plasma membrane. The relocalization of the 5-phosphatase resulted in a sharp reduction in PI(4,5)P2 and cortical actin levels. I therefore went on to characterize the effects of PI(4,5)P2 and actin depletion on cell contractility during ventral furrow formation. Illumination of the entire embryo with a 488 nm (single photon) laser resulted in the inhibition of both apical constriction and tissue folding. After this encouraging result, I sought to achieve spatial specificity in addition to temporal precision by photo-activating smaller groups of cells. However, light scattering within the tissue resulted in unwanted photo-activation of cells neighboring the illuminated areas, making the single photon approach impracticable. By discussing with in-house microscopy experts, I found out that this limitation could be overcome by using two-photon microscopy. Indeed, the requirement for near-simultaneous absorption of two photons makes it very likely that photo-activation remains limited to the area where the laser is focused. I therefore set up a protocol to achieve optimal levels of photo-activation with a two-photon laser (950 nm). To my delight, photo-activation could be achieved at the seconds time-scale and allowed high spatial precision, namely single cell resolution.
Having established this powerful approach, I tackled two interesting questions regarding ventral furrow formation. In particular, I tested whether apical constriction in ventral cells is required only to initiate the process of tissue bending, or whether it is necessary throughout the process to achieve complete folding of the tissue. By modulating apical constriction only in ventral cells, I could show that apical constriction is necessary not only to kick off, but also to sustain tissue folding.
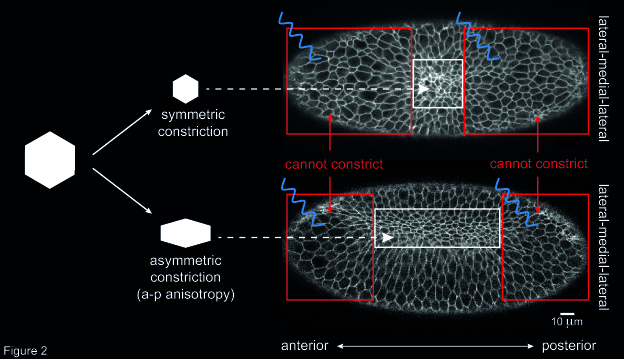
Another open question concerned the way individual cells constrict during ventral furrow formation. When ventral cells reduce their apical surface, they do so in an asymmetric fashion: cells shrink along the embryo’s medial-lateral axis and remain elongated along the anterior-posterior (a-p) axis. This asymmetry in cell constriction, known as a-p anisotropy, is thought to be the result of higher tension along the a-p axis than along the medial-lateral axis. However, the origin of this tension was unknown. To address this question, I altered the geometry of the primordium by inhibiting cell contractility in two areas at the anterior and posterior end of the ventral furrow tissue. Then, I checked whether this resulted in any change in a-p anisotropy in cells that were left able to constrict (i.e. not illuminated with blue light). I could show that the degree of a-p anisotropy in non-illuminated cells was higher if the two illuminated areas were further apart than if they were close together along the a-p axis (Figure 2). In other words, if the rectangular geometry of the constricting tissue was preserved cells constricted in an asymmetric way. Instead, if the constricting patch of cells was squared-shaped, constriction was more symmetric, and resulted in roundish cells. This suggests that the geometry of the ventral furrow tissue impacts on the way individual cells constrict.
As I collected increasing amount of data, I needed to transform a lot of visual information into meaningful numbers, so I teamed up with Joseph Barry, who at that time was a postdoc in Wolfgang Huber’s group at EMBL. Joe did a great job of developing algorithms to quantify cell features, such as area and a-p anisotropy. For this project, I heavily relied on the EMBL Advanced Light Microscopy Facility, the state-of-the-art equipment available in the Developmental Biology Unit, and of course the valuable input of my supervisor Stefano De Renzis. Shortly after publication in Developmental Cell14, our optogenetic approach generated the interest of many research groups around the world, as apical constriction is a highly conserved cellular mechanism during morphogenesis. However, besides apical constriction, cell contractility drives a multitude of cell behaviors, from cytokinesis to cell migration. Therefore, I expect that our approach will be useful to the community for addressing intriguing questions about animal development.
In the meanwhile, optogenetics has enabled the modulation of many other cellular activities, including organelle transport, cell-cell signalling, apoptosis, and gene expression15-20. The translation of these tools to living organisms will undoubtedly provide new insights into the mechanisms by which embryos grow and develop. As a developmental biologist and recent PhD graduate, I am very much looking forward to seeing where lights will guide us.
1. Method of the Year 2010. Nat Meth 8, 1-1, doi:10.1038/nmeth.f.321 (2011). ↩
2. Insights of the decade. Stepping away from the trees for a look at the forest. Introduction. Science (New York, N.Y.) 330, 1612-1613, doi:10.1126/science.330.6011.1612 (2010). ↩
3. Zemelman, B. V., Lee, G. A., Ng, M. & Miesenbock, G. Selective photostimulation of genetically chARGed neurons. Neuron 33, 15-22 (2002). ↩
4. Banghart, M., Borges, K., Isacoff, E., Trauner, D. & Kramer, R. H. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci 7, 1381-1386, doi:10.1038/nn1356 (2004). ↩
5. Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8, 1263-1268 (2005). ↩
6. Wu, Y. I. et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104-108, doi:10.1038/nature08241 (2009). ↩
7. Wang, X., He, L., Wu, Y. I., Hahn, K. M. & Montell, D. J. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nature cell biology 12, 591-597, doi:10.1038/ncb2061 (2010). ↩
8. Toettcher, J. E., Gong, D., Lim, W. A. & Weiner, O. D. Light-based feedback for controlling intracellular signaling dynamics. Nature methods 8, 837-839, doi:10.1038/nmeth.1700 (2011). ↩
9. Reversi, A., Loeser, E., Subramanian, D., Schultz, C. & De Renzis, S. Plasma membrane phosphoinositide balance regulates cell shape during Drosophila embryo morphogenesis. The Journal of cell biology 205, 395-408, doi:10.1083/jcb.201309079 (2014). ↩
10. Levskaya, A., Weiner, O. D., Lim, W. A. & Voigt, C. A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997-1001, doi:10.1038/nature08446 (2009).↩
11. Yazawa, M., Sadaghiani, A. M., Hsueh, B. & Dolmetsch, R. E. Induction of protein-protein interactions in live cells using light. Nature biotechnology 27, 941-945, doi:10.1038/nbt.1569 (2009).↩
12. Kennedy, M. J. et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nature methods 7, 973-975, doi:10.1038/nmeth.1524 (2010).↩
13. Idevall-Hagren, O., Dickson, E. J., Hille, B., Toomre, D. K. & De Camilli, P. Optogenetic control of phosphoinositide metabolism. Proceedings of the National Academy of Sciences of the United States of America 109, E2316-2323, doi:10.1073/pnas.1211305109 (2012).↩
14. Guglielmi, G., Barry, J. D., Huber, W. & De Renzis, S. An Optogenetic Method to Modulate Cell Contractility during Tissue Morphogenesis. Developmental cell, doi:10.1016/j.devcel.2015.10.020 (2015).↩
15. Mills, E., Chen, X., Pham, E., Wong, S. & Truong, K. Engineering a photoactivated caspase-7 for rapid induction of apoptosis. ACS synthetic biology 1, 75-82, doi:10.1021/sb200008j (2012). ↩
16. Liu, H., Gomez, G., Lin, S., Lin, S. & Lin, C. Optogenetic control of transcription in zebrafish. PloS one 7, e50738, doi:10.1371/journal.pone.0050738 (2012).↩
17. Toettcher, J. E., Weiner, O. D. & Lim, W. A. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422-1434, doi:10.1016/j.cell.2013.11.004 (2013).↩
18. Grusch, M. et al. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. The EMBO journal 33, 1713-1726, doi:10.15252/embj.201387695 (2014).↩
19. Motta-Mena, L. B. et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nature chemical biology 10, 196-202, doi:10.1038/nchembio.1430 (2014).↩
20. van Bergeijk, P., Adrian, M., Hoogenraad, C. C. & Kapitein, L. C. Optogenetic control of organelle transport and positioning. Nature 518, 111-114, doi:10.1038/nature14128 (2015).↩
 (5 votes)
(5 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)
 (4 votes)
(4 votes)