The 4th meeting in the Abcam Adult Neurogenesis conference series was held in the beautiful city of Dresden earlier this year. The conference’s aim was to put the developmental process of adult neurogenesis and its regulation into the wider context of its functional and presumed evolutionary relevance.
The 4th meeting in the Abcam Adult Neurogenesis conference series was held in the beautiful city of Dresden earlier this year. The conference’s aim was to put the developmental process of adult neurogenesis and its regulation into the wider context of its functional and presumed evolutionary relevance.
Reporting from the meeting was our roving reporter, Nambirajan Govindarajan (winner of The Node and Abcam’s joint Meeting Reporter competition). He tweeted his way through the meeting and has written an insightful report of this meeting. Thanks Govind!
Adult Neurogenesis at 50: the Dresden chronicles
This year marks the 50th anniversary of the first publication showing adult neurogenesis in mammals (Altman and Das, 1965). The growth and development of this field was commemorated by the Abcam conference on Adult Neurogenesis: Evolution, Regulation and Function in May this year in the beautiful baroque city of Dresden.
All good tales start with “Once upon a time,” and this was no exception. Gerd Kempermann kickstarted the meeting with his enchanting narrative on the serendipitous birth of the field and its ebbs and flows over the past five decades. While studying neuronal activity in the adult rat brain, Joseph Altman switched from tritiated leucine to the more sensitive tritiated thymidine to calibrate his detection and accidentally observed proliferating cells in the rat hippocampus. He published his findings with Gopal Das in a seminal paper, which gave birth to the ‘science of the future’ that would change Santiago Ramón y Cajal’s harsh decree (Ramón y Cajal, 1928; Altman and Das, 1965). This chance discovery has spawned a long line of research on adult neurogenesis in many avian, piscine and mammalian species. Kempermann recounted various contrasts and conflicts among the research and researchers in the field and even dispelled some outstanding myths. He emphasised the role of adult neurogenesis in mediating brain plasticity, which correlates with neural complexity along the evolutionary tree. Being the expert raconteur, Kempermann delighted us all and set the stage for the meeting.
Open questions in adult neurogenesis
The eminent Fred Gage laid forth the big questions facing adult neurogenesis in his keynote lecture. He focused on the role of the niche in regulating neurogenesis. How different are the two known neurogenic zones – the subventricular zone (SVZ) and the dentate gyrus (DG)? How are the precursors affected by the niche vasculature and metabolic state? Taking it a step further, Gage even wondered if a cell isolated from a non-neurogenic area would become neurogenic if transplanted into either niche. Tackling these questions is essential in understanding the function, regulation and evolution of adult neurogenesis. Gage also remarked that other potential niches such as the striatum, angular gyrus, cortex and spinal cord need to be investigated. He then reminded us of the standing questions in the field – the precise molecular nature of different neurogenic cell types and the comparability of the in vitro and in vivo scenarios. It is also unclear how the number of divisions of a precursor cell is regulated. Gage also emphasised the need to understand the integration of newborn neurons in the neural network and whether cellular excitability and network activity influence this process. Another heavily debated issue is whether and how adult neurogenesis contributes to cognition. The mechanisms that modulate environmental regulation of adult neurogenesis are still unknown. These are all open questions that Gage urged us to pursue and discuss. Finally, anticipating the biggest question on everyone’s mind, Gage also brought up the role of adult neurogenesis in ageing and neurological disease. Can restoring adult neurogenesis help a degenerating brain? These open questions promise to fuel neurogenesis research further and hopefully unravel the intricate codes governing this enigmatic part of us we call our brain.
Born in the wild
Neurons are born in the wild too! To enrich laboratory research with some wild data, this meeting brought together some species very different from inbred rodents – mole-rats, silver foxes, dolphins and warblers. Irmgard Amrein introduced a technical issue that we lab folks take for granted. How does one assess a wild animal’s age? Amrein has used parameters such as teeth wear, lens weight and bone maturation to closely estimate age in the wild. Her group has found that adult neurogenesis is lower in subterranean rodent species compared to terrestrial ones (Amrein et al., 2014). According to Amrein, lower habitat demand on subterranean species correlates with lower adult hippocampal neurogenesis (AHN). She then moved on to the effect of domestication and social interaction on AHN in wild and farm-bred silves foxes at Dimitri K. Belyaev’s unique fox farm in Novosibirsk. Belyaev has selectively bred silver foxes (Vulpes vulpes) based on their tameness, which he believes is how they were originally domesticated (Belyaev, 1979). These domesticated foxes selected for their tameness showed higher hippocampal cell proliferation and neuronal differentiation compared to unselected controls (Huang et al., 2015). These findings suggest that AHN might be involved in interspecific social interaction. We then dived into the cetaecean brain with Paul Manger who vivdly illustrated that dolphins lack AHN, which correlates with their aquatic habitat, lack of olfaction underwater and early-life insomnia. Even as grown-ups, dolphins sleep without any REM phase and possess a small, rudimentary DG with almost no AHN (Patzke et al., 2015). We need to test this interesting hypothesis in other sleepless species before firmly establishing a link. Manger also found that doublecortin immunoreactivity rapidly decreased with post-mortem delay and thus rendered the ex vivo analysis of neuronal differentiation in wild species extremely challenging. From the oceans Anat Barnea took us flying with her migrant reed warbler (Acrocephalus scirpaceous) that recruits more new neurons than the resident, closely-related Clamorous warbler (A. stentoreus) (Barkan et al., 2014). Barnea’s findings support the role of adult neurogenesis in spatial navigation and adaptation to changing environments. Altogether, the ‘wild’ talks gave us an insight into how adult neurogenesis could have evolved and how it affects the behaviour of animals in their natural habitat.
Studied in the lab
We moved from the wilderness to the laboratory. Wieland Huttner touched upon the evolutionary expansion of the human neocortex, which results from embryonic basal progenitor cell proliferation in the SVZ. Huttner presented the role of ARHGAP11B, a gene that was partially duplicated after the human evolutionary lineage split from the chimpanzee. This gene promotes basal progenitor generation and proliferation in mice, can induce folding of the developing mouse neocortex, and might have contributed to the expansion of the human neocortex (Florio et al., 2015). We delved deeper into the molecular mechanisms regulating neurogenesis. Federico Calegari presented his unique approach combining DNA adenine methyltransferase identification (DamID) with deep sequencing to discover the function of novel genes involved in corticogenesis (Aprea et al., 2013). His group has characterised Tox, a novel switch gene that regulates cortical development in mice (Artegiani et al., 2015). Stephan Schwarzacher discussed the integration of newborn neurons. By studying the activation of immediate early genes including c-Fos, Arc and Zif after high-frequency stimulation, Schwarzacher has concluded that full functional integration of newborn neurons follows and closely correlates with their structural maturation (Jungenitz et al., 2014). Juan M. Encinas has stimulated the hippocampus with kainic acid (KA) and observed that seizures induced neural stem cells (NSCs) to form reactive astrocytes whereas subthreshold excitation activated the NSCs and eventually triggered them to form astrocytes. In both cases, Encinas found that KA impaired neurogenesis in the long term (Sierra et al., 2015). Live imaging of newborn neurons in vivo is one of the technological breakthroughs of the 21st century. Fred Gage and Sebastian Jessberger both presented this approach to investigate AHN and dendritic morphology of newborn neurons. Jessberger impressed us with the depth of imaging achieved, using a cranial window preparation that leaves the hippocampal formation intact, including the CA1. Hongjun Song presented his findings on distinct radial glia-like stem cell populations, labelled by specific markers such as Nestin, Gli and Mash, and discussed their properties. Song’s pioneering work on single-cell analysis of adult neurogenesis is sure to pave the way in analysing specific cell populations and unravelling the cellular heterogeneity in the neurogenic niches.
Learning what they do
Studying the evolution and regulation of adult neurogenesis leads us to the most intriguing question: how is adult neurogenesis involved in brain function? Paul Frankland refreshed our memory of the role of AHN in forgetting. According to Frankland, upregulating AHN by running induces forgetting in mice (Akers et al., 2014). Interestingly, Frankland has found that during reversal learning in the water maze, mice seem to learn the new location of the platform better with more new neurons in their hippocampi. But does that mean they have forgotten where the platform was previously placed? Frankland’s findings sparked an exciting discussion on the nature of memory and how the brain actually forgets a memory. Spatial learning was further discussed by Nora Abrous whose recent work suggests that water maze learning does not affect cell proliferation or survival in the adult mouse DG (Trinchero et al., 2015). Benedikt Berninger discussed a different angle on the role of experience in regulating adult neurogenesis. Berninger reported that environmental enrichment within a critical window of 2-6 weeks after the birth of new neurons significantly modulates their development and integration (Bergami et al., 2015).
The relation between AHN and stress was keenly discussed at the meeting. Carlos Fitzsimons ‘stressed’ that glucocorticoids regulate stress-induced adult neurogenesis through epigenetic mechanisms. Corticosterone treatment transiently reduced proliferation by modulating DNA methylation. Could this modulate the inheritance of stress effects? Recent work by Fitzsimons demonstrates that the glucocorticoid receptor regulates the maturation and integration of newborn neurons and plays an important role in contextual fear conditioning (Fitzsimons et al., 2013). Another interesting talk along these lines by Yassemi Koutmani discussed the role of corticotrophin-releasing hormone (CRH) in upregulating AHN thereby reversing the damage by glucocorticoid treatment. Koutmani’s work depicts that CRHR1 is critically involved in regulating how NSCs respond to environmental stimuli (Koutmani et al., 2013). Friederike Klempin presented her findings on the role of ACE2 activity that sustains brain serotonin level, which in turn mediates the fast neurogenic response of the niche to physical activity (Klempin et al., 2013).
When it comes to regenerating the brain, the zebrafish swims miles ahead of mammals. Caghan Kizil has generated a zebrafish model for chronic neurodegeneration by injecting Aß42 peptides into their brain, which leads to cell death, inflammation, synaptic degeneration and memory impairment. However, Kizil found that unlike in mammals, Aß42 treatment triggered stem cells in the zebrafish brain to proliferate and remarkably form new neurons. Such studies on neuroregeneration in the fish can be extremely valuable in designing new therapies against neurodegeneration.
Concluding remarks
Half a century bygone and adult neurogenesis has evolved into a mainstream research discipline in neurobiology. It was born by chance, differentiated into a distinct lineage, carved out its own niche, migrated all over the world and has integrated perfectly within the extensive network of biomedical science, mirroring the life of the very cells it studies. Many questions have been answered and many more daunt us still. With further technical innnovations, better models and cross-species experiments, the coming decades are bound to keep us busy uncovering the secrets of how and why our brains make new neurons lifelong.
References
Akers, K. G., Martinez-Canabal, A., Restivo, L., Yiu, A. P., De Cristofaro, A., Hsiang, H. L., Wheeler, A. L., Guskjolen, A., Niibori, Y., Shoji, H. et al. (2014) ‘Hippocampal neurogenesis regulates forgetting during adulthood and infancy’, Science 344(6184): 598-602.
Altman, J. and Das, G. D. (1965) ‘Post-natal origin of microneurones in the rat brain’, Nature 207(5000): 953-6.
Amrein, I., Becker, A. S., Engler, S., Huang, S. H., Muller, J., Slomianka, L. and Oosthuizen, M. K. (2014) ‘Adult neurogenesis and its anatomical context in the hippocampus of three mole-rat species’, Frontiers in neuroanatomy 8: 39.
Aprea, J., Prenninger, S., Dori, M., Ghosh, T., Monasor, L. S., Wessendorf, E., Zocher, S., Massalini, S., Alexopoulou, D., Lesche, M. et al. (2013) ‘Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment’, The EMBO journal 32(24): 3145-60.
Artegiani, B., de Jesus Domingues, A. M., Bragado Alonso, S., Brandl, E., Massalini, S., Dahl, A. and Calegari, F. (2015) ‘Tox: a multifunctional transcription factor and novel regulator of mammalian corticogenesis’, The EMBO journal 34(7): 896-910.
Barkan, S., Yom-Tov, Y. and Barnea, A. (2014) ‘A possible relation between new neuronal recruitment and migratory behavior in Acrocephalus warblers’, Developmental neurobiology 74(12): 1194-209.
Belyaev, D. K. (1979) ‘The Wilhelmine E. Key 1978 invitational lecture. Destabilizing selection as a factor in domestication’, The Journal of heredity 70(5): 301-8.
Bergami, M., Masserdotti, G., Temprana, S. G., Motori, E., Eriksson, T. M., Gobel, J., Yang, S. M., Conzelmann, K. K., Schinder, A. F., Gotz, M. et al. (2015) ‘A critical period for experience-dependent remodeling of adult-born neuron connectivity’, Neuron 85(4): 710-7.
Fitzsimons, C. P., van Hooijdonk, L. W., Schouten, M., Zalachoras, I., Brinks, V., Zheng, T., Schouten, T. G., Saaltink, D. J., Dijkmans, T., Steindler, D. A. et al. (2013) ‘Knockdown of the glucocorticoid receptor alters functional integration of newborn neurons in the adult hippocampus and impairs fear-motivated behavior’, Molecular psychiatry 18(9): 993-1005.
Florio, M., Albert, M., Taverna, E., Namba, T., Brandl, H., Lewitus, E., Haffner, C., Sykes, A., Wong, F. K., Peters, J. et al. (2015) ‘Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion’, Science 347(6229): 1465-70.
Huang, S., Slomianka, L., Farmer, A. J., Kharlamova, A. V., Gulevich, R. G., Herbeck, Y. E., Trut, L. N., Wolfer, D. P. and Amrein, I. (2015) ‘Selection for tameness, a key behavioral trait of domestication, increases adult hippocampal neurogenesis in foxes’, Hippocampus.
Jungenitz, T., Radic, T., Jedlicka, P. and Schwarzacher, S. W. (2014) ‘High-frequency stimulation induces gradual immediate early gene expression in maturing adult-generated hippocampal granule cells’, Cerebral cortex 24(7): 1845-57.
Klempin, F., Beis, D., Mosienko, V., Kempermann, G., Bader, M. and Alenina, N. (2013) ‘Serotonin is required for exercise-induced adult hippocampal neurogenesis’, The Journal of neuroscience : the official journal of the Society for Neuroscience 33(19): 8270-5.
Koutmani, Y., Politis, P. K., Elkouris, M., Agrogiannis, G., Kemerli, M., Patsouris, E., Remboutsika, E. and Karalis, K. P. (2013) ‘Corticotropin-releasing hormone exerts direct effects on neuronal progenitor cells: implications for neuroprotection’, Molecular psychiatry 18(3): 300-7.
Patzke, N., Spocter, M. A., Karlsson, K. A. E., Bertelsen, M. F., Haagensen, M., Chawana, R., Streicher, S., Kaswera, C., Gilissen, E., Alagaili, A. N. et al. (2015) ‘In contrast to many other mammals, cetaceans have relatively small hippocampi that appear to lack adult neurogenesis’, Brain structure & function 220(1): 361-83.
Ramón y Cajal, S. (1928) Degeneration and Regeneration of the Nervous System: Oxford Univ. Press, London.
Sierra, A., Martin-Suarez, S., Valcarcel-Martin, R., Pascual-Brazo, J., Aelvoet, S. A., Abiega, O., Deudero, J. J., Brewster, A. L., Bernales, I., Anderson, A. E. et al. (2015) ‘Neuronal hyperactivity accelerates depletion of neural stem cells and impairs hippocampal neurogenesis’, Cell stem cell 16(5): 488-503.
Trinchero, M. F., Koehl, M., Bechakra, M., Delage, P., Charrier, V., Grosjean, N., Ladeveze, E., Schinder, A. F. and Abrous, D. N. (2015) ‘Effects of spaced learning in the water maze on development of dentate granule cells generated in adult mice’, Hippocampus.
 (4 votes)
(4 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet) (3 votes)
(3 votes)







 We recently interviewed Brigid Hogan, a developmental biologist who has worked extensively on the early stages of mouse development and is now unravelling the mysteries of lung organogenesis. She is the George Barth Geller Professor and Chair of the Department of Cell Biology at Duke University Medical Center. Brigid is also the winner of the 2015 Society for Developmental Biology (SDB) Lifetime Achievement Award. See the Spotlight article on p.
We recently interviewed Brigid Hogan, a developmental biologist who has worked extensively on the early stages of mouse development and is now unravelling the mysteries of lung organogenesis. She is the George Barth Geller Professor and Chair of the Department of Cell Biology at Duke University Medical Center. Brigid is also the winner of the 2015 Society for Developmental Biology (SDB) Lifetime Achievement Award. See the Spotlight article on p.  The retromer complex is a multimeric protein complex involved in recycling proteins from endosomes to the trans-Golgi network or plasma membrane. Here, Wang and Bellen summarise the role of the retromer complex in developmental processes, neuronal maintenance, and human neurodegenerative diseases. See the Development at a Glance article on p.
The retromer complex is a multimeric protein complex involved in recycling proteins from endosomes to the trans-Golgi network or plasma membrane. Here, Wang and Bellen summarise the role of the retromer complex in developmental processes, neuronal maintenance, and human neurodegenerative diseases. See the Development at a Glance article on p. 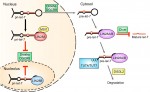 LIN28 is an RNA-binding protein best known for its roles in promoting pluripotency via regulation of the microRNA let-7. However, recent studies have uncovered new roles for LIN28, suggesting that it is more than just a regulator of miRNA biogenesis. Here, Tsialikas and Romer-Seibert review how LIN28 functions in development and disease. See the Primer on p.
LIN28 is an RNA-binding protein best known for its roles in promoting pluripotency via regulation of the microRNA let-7. However, recent studies have uncovered new roles for LIN28, suggesting that it is more than just a regulator of miRNA biogenesis. Here, Tsialikas and Romer-Seibert review how LIN28 functions in development and disease. See the Primer on p.  The 4th meeting in the Abcam Adult Neurogenesis conference series was held in the beautiful city of Dresden earlier this year. The conference’s aim was to put the developmental process of adult neurogenesis and its regulation into the wider context of its functional and presumed evolutionary relevance.
The 4th meeting in the Abcam Adult Neurogenesis conference series was held in the beautiful city of Dresden earlier this year. The conference’s aim was to put the developmental process of adult neurogenesis and its regulation into the wider context of its functional and presumed evolutionary relevance.