Here is some developmental biology related content from other journals published by The Company of Biologists.

Drosophila helps to elucidate the effects of radiation therapy
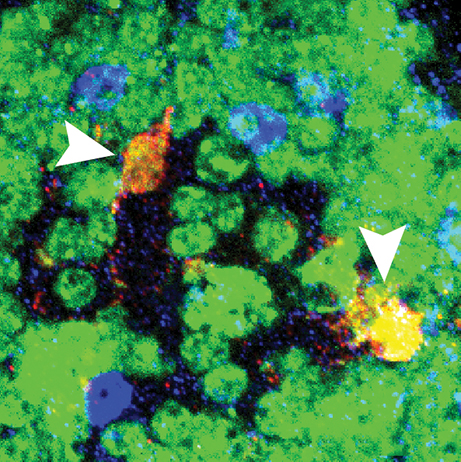 Children who are exposed to cranial radiation therapy (CRT) to treat brain tumours are at increased risk later in life for neurocognitive, motor and seizure disorders. Current radioprotective agents are difficult to administer and have severe side effects. Thus, Barry Ganetzky and colleagues used Drosophila to model and elucidate the toxic effects of CRT during juvenile development. First, they found that irradiating fly larvae with different doses of radiation reduces the percentage of animals that survive to adulthood and impairs motor activity of surviving adults, in a dose-dependent manner. These phenotypes are paralleled by an increased expression of activated death caspase-1, a cell death marker, in adult brains. Then, using flies at different developmental stages, they found that the toxic effects of radiation exposure become more severe with decreasing larval age at time of irradiation. Notably, flies with different genetic backgrounds show different sensitivity to radiation. Thus, Drosophila represents a useful system to model CRT-induced toxicity and to dissect key molecular and genetic pathways. These, in turn, might illuminate potential targets for radioprotective purposes. Read the paper here (Open Access).
Children who are exposed to cranial radiation therapy (CRT) to treat brain tumours are at increased risk later in life for neurocognitive, motor and seizure disorders. Current radioprotective agents are difficult to administer and have severe side effects. Thus, Barry Ganetzky and colleagues used Drosophila to model and elucidate the toxic effects of CRT during juvenile development. First, they found that irradiating fly larvae with different doses of radiation reduces the percentage of animals that survive to adulthood and impairs motor activity of surviving adults, in a dose-dependent manner. These phenotypes are paralleled by an increased expression of activated death caspase-1, a cell death marker, in adult brains. Then, using flies at different developmental stages, they found that the toxic effects of radiation exposure become more severe with decreasing larval age at time of irradiation. Notably, flies with different genetic backgrounds show different sensitivity to radiation. Thus, Drosophila represents a useful system to model CRT-induced toxicity and to dissect key molecular and genetic pathways. These, in turn, might illuminate potential targets for radioprotective purposes. Read the paper here (Open Access).

Of oocytes and PDKs
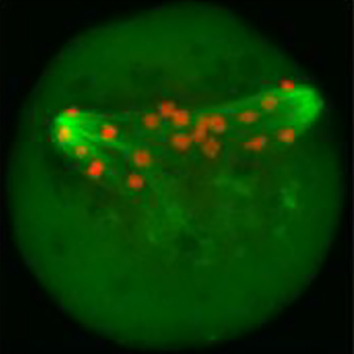 Perturbations in energy metabolism cause meiotic defects in mammalian oocytes. It has been suggested that these may be exerted through pyruvate dehydrogenase kinases (PDKs) that act on the pyruvate dehydrogenase complex (PDH), which, in somatic cells, is inhibited by phosphorylation of its catalytic subunit PDHE1α at serine residues S232, S292 and S300; however, the role of particular PDKs in the maturation of meiotic oocytes is still unclear. Qiang Wang and colleagues, therefore, examined the individual contributions of known mammalian PDKs to the regulation of murine oocyte maturation. They found that oocytes that overexpress PDK3 have elevated levels of PDHE1α phosphorylated at S293, defects in polar body 1 (Pb1) extrusion, as well as a disorganised spindle and misaligned chromosomes. Moreover, in keeping with a central role of PDH in energy production, the authors also found decreased levels of ATP. These spindle and chromosome abnormalities, as well as metabolic defects could be rescued with a phosphorylation resistant PDH-S293A mutant. By contrast, knockdown of PDK1 or PDK2, both of which phosphorylate S232, caused defects in Pb1 extrusion, spindle assembly and chromosome alignment, but did not affect the level of total ATP. Taken together, these results reveal that PDK3-dependent phosphorylation of PDHE1α at S293 affects oocyte maturation through PDH inhibition and changes in metabolism, whereas PDK1/PDK2-mediated PDHE1α phosphorylation at S232 has effects on oocyte meiosis that may be independent of PDH’s role in energy production. Read the paper here .
Perturbations in energy metabolism cause meiotic defects in mammalian oocytes. It has been suggested that these may be exerted through pyruvate dehydrogenase kinases (PDKs) that act on the pyruvate dehydrogenase complex (PDH), which, in somatic cells, is inhibited by phosphorylation of its catalytic subunit PDHE1α at serine residues S232, S292 and S300; however, the role of particular PDKs in the maturation of meiotic oocytes is still unclear. Qiang Wang and colleagues, therefore, examined the individual contributions of known mammalian PDKs to the regulation of murine oocyte maturation. They found that oocytes that overexpress PDK3 have elevated levels of PDHE1α phosphorylated at S293, defects in polar body 1 (Pb1) extrusion, as well as a disorganised spindle and misaligned chromosomes. Moreover, in keeping with a central role of PDH in energy production, the authors also found decreased levels of ATP. These spindle and chromosome abnormalities, as well as metabolic defects could be rescued with a phosphorylation resistant PDH-S293A mutant. By contrast, knockdown of PDK1 or PDK2, both of which phosphorylate S232, caused defects in Pb1 extrusion, spindle assembly and chromosome alignment, but did not affect the level of total ATP. Taken together, these results reveal that PDK3-dependent phosphorylation of PDHE1α at S293 affects oocyte maturation through PDH inhibition and changes in metabolism, whereas PDK1/PDK2-mediated PDHE1α phosphorylation at S232 has effects on oocyte meiosis that may be independent of PDH’s role in energy production. Read the paper here .
Fly centrioles – different after all
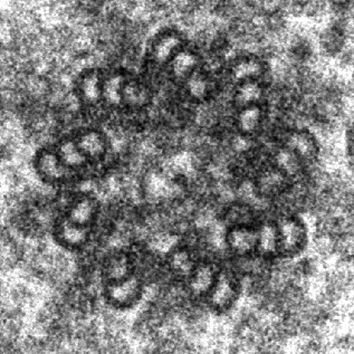 In contrast to vertebrate centriole pairs, where the mother and daughter centriole can easily be distinguished from each other by morphology, Drosophila centrioles lack structural differences and can only be recognised by identifying specific protein markers. Marco Gottardo, Giuliano Callaini and Maria Giovanna Riparbelli used electron microscopy to examine mother and daughter centrioles in a range of fly tissues. They found that mature centrioles of somatic cells comprised doublets with a distinct cartwheel structure, whereas differentiating male germ cells had centrioles that comprised triplets. However, no structural differences between the mother and daughter centrioles could be found in any of the tissues examined. By contrast, male germline stem cells (GSCs) contained centrioles that comprised doublets, triplets or a mix thereof. Upon careful examination of the GSC niche, the authors observed that the mother centriole comprised triplets and that the daughter centriole was made up of doublets that matured into triplets. In addition, the authors showed that the mother centriole extended projections to the apical plasma membrane, which could function as an anchor to keep the mother centriole stationary during centrosome separation. This study is the first demonstration of a structural asymmetry between mother and daughter centrioles in Drosophila, which might reflect an unequal distribution of centrosomal proteins, thereby providing important insights into centrosome biology in flies. Read the paper here .
In contrast to vertebrate centriole pairs, where the mother and daughter centriole can easily be distinguished from each other by morphology, Drosophila centrioles lack structural differences and can only be recognised by identifying specific protein markers. Marco Gottardo, Giuliano Callaini and Maria Giovanna Riparbelli used electron microscopy to examine mother and daughter centrioles in a range of fly tissues. They found that mature centrioles of somatic cells comprised doublets with a distinct cartwheel structure, whereas differentiating male germ cells had centrioles that comprised triplets. However, no structural differences between the mother and daughter centrioles could be found in any of the tissues examined. By contrast, male germline stem cells (GSCs) contained centrioles that comprised doublets, triplets or a mix thereof. Upon careful examination of the GSC niche, the authors observed that the mother centriole comprised triplets and that the daughter centriole was made up of doublets that matured into triplets. In addition, the authors showed that the mother centriole extended projections to the apical plasma membrane, which could function as an anchor to keep the mother centriole stationary during centrosome separation. This study is the first demonstration of a structural asymmetry between mother and daughter centrioles in Drosophila, which might reflect an unequal distribution of centrosomal proteins, thereby providing important insights into centrosome biology in flies. Read the paper here .
New roles for ROPs in plant growth
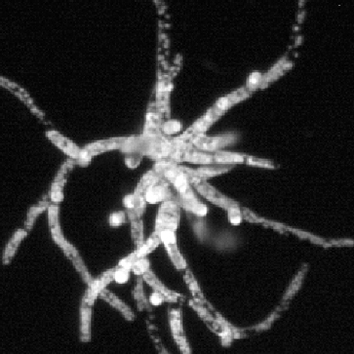 Rho of plant (ROP) proteins belong to the sole family of Rho-like GTPases in plants and have been suggested to regulate the polarised growth of tip-growing cells by modulating interactions between the actin and microtubule cytoskeletons. However, most studies performed to date have relied on the overexpression of proteins, including constitutively active or dominant-negative isoforms, and therefore Magdalena Bezanilla and colleagues took advantage of the ease of reverse genetics and the low number of highly similar ROP proteins (ROP1–4) in the model moss Physcomitrella patens to analyse the role of ROPs using a loss-of-function approach. Knockdown of all four ROP genes by RNAi resulted in dwarf plants comprising small spherical cells that had a significantly smaller area, compared with control plants. The authors also observed that, surprisingly, ROP-knockdown plants had strongly reduced cell adhesion and an altered cell wall structure, suggesting that ROPs might have previously unappreciated roles. Importantly, the authors found that actin filaments were more dynamic and that actin arrays were more disordered in ROP-knockdown cells, but that there was no ROP-specific effect on the dynamics of the microtubule cytoskeleton. In summary, this extensive study provides important new insights into the functions of ROP proteins in cell adhesion, cell wall assembly and polarised cell growth – roles they possibly exert by suppressing actin dynamics. Read the paper here .
Rho of plant (ROP) proteins belong to the sole family of Rho-like GTPases in plants and have been suggested to regulate the polarised growth of tip-growing cells by modulating interactions between the actin and microtubule cytoskeletons. However, most studies performed to date have relied on the overexpression of proteins, including constitutively active or dominant-negative isoforms, and therefore Magdalena Bezanilla and colleagues took advantage of the ease of reverse genetics and the low number of highly similar ROP proteins (ROP1–4) in the model moss Physcomitrella patens to analyse the role of ROPs using a loss-of-function approach. Knockdown of all four ROP genes by RNAi resulted in dwarf plants comprising small spherical cells that had a significantly smaller area, compared with control plants. The authors also observed that, surprisingly, ROP-knockdown plants had strongly reduced cell adhesion and an altered cell wall structure, suggesting that ROPs might have previously unappreciated roles. Importantly, the authors found that actin filaments were more dynamic and that actin arrays were more disordered in ROP-knockdown cells, but that there was no ROP-specific effect on the dynamics of the microtubule cytoskeleton. In summary, this extensive study provides important new insights into the functions of ROP proteins in cell adhesion, cell wall assembly and polarised cell growth – roles they possibly exert by suppressing actin dynamics. Read the paper here .

Consequences of calcium decline on the embryogenesis and life history of Daphnia magna
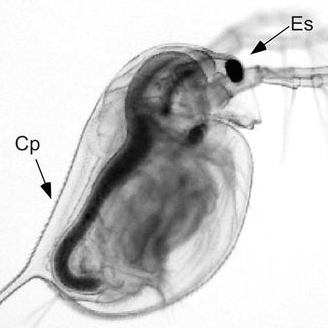 Ambient calcium is declining in thousands of soft-water lake habitats. Daphnia females provision their offspring with calcium and through that incur a potential allocation trade-off. Read the paper here.
Ambient calcium is declining in thousands of soft-water lake habitats. Daphnia females provision their offspring with calcium and through that incur a potential allocation trade-off. Read the paper here.
Diverse dose–response effects of yolk androgens on embryo development and nestling growth in a wild passerine
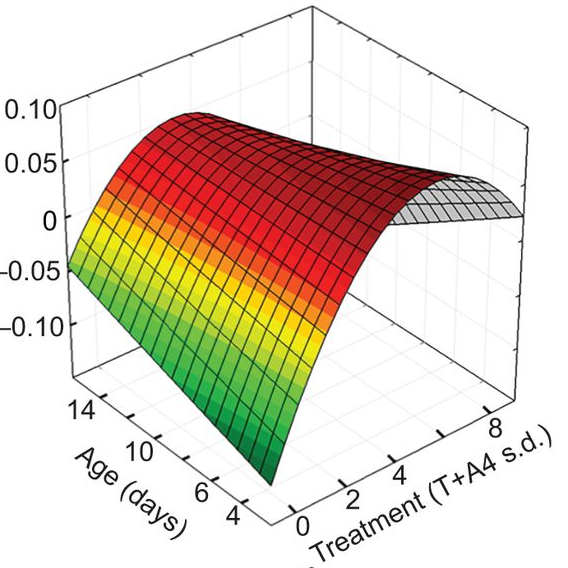 Yolk androgens in wild passerines show complex dose–response effects during early development, including both linear and non-linear responses for different traits. Read the paper here.
Yolk androgens in wild passerines show complex dose–response effects during early development, including both linear and non-linear responses for different traits. Read the paper here.
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet) (23 votes)
(23 votes)


 (3 votes)
(3 votes)







