This is the final post from our developmental neurobiology seminar this semester. Two students wrote about our discussion of the importance of neuronal activity during synaptogenesis and their professor combined and edited the pieces. As usual, we focused on development in the vertebrate retina. Hope you’ve enjoyed our contributions, we’ve enjoyed sharing our new-found understanding.
“Use it or lose it.” One of my mother’s favorite phrases, applicable to any number of situations. Fail to practice piano? Don’t use a toy for a while? Realize you’ve forgotten your high school Spanish? “Use it or lose it.”
The concept of “use it or lose it” can also be applied to synaptogenesis and neural circuit development (although I doubt my mother was thinking of this when she used the phrase). The idea of Hebbian connections – that synapse formation and elimination is dependent on activity – has been primarily supported by studies of the neuromuscular junction [1]. In general, input neurons converging on dendrites differ in synaptic activity, resulting in more active inputs outcompeting less active inputs for dendrite connection. Inputs with less active synaptic regions are ultimately eliminated [2],[3]. Essentially, the less active neurons don’t “use it” and so they “lose it”, with “it” being an electrochemical connection to a neighboring neuron.
Several years ago, investigations of developing connections in the vertebrate retina suggested that this idea of “use it or lose it” might not be the sole mechanism employed within the developing nervous system. Our class recently read and discussed a paper by Kerschensteiner et al. (2009) [4]. This detailed study suggested that, in contrast to the classical model, some neuronal connections are formed and maintained in the inner plexiform layer independently of synaptic activity.
Anatomy and Physiology Background: ON and OFF cells in the Inner Plexiform Layer
When light hits the mammalian eye, it changes the electrical properties of photoreceptors so that their downstream partners, bipolar cells, deliver a message to retinal ganglion cells (RGCs), which in turn relay information about visual cues to the brain (see Figure 1). The middlemen in this process, bipolar cells, can be depolarized when light hits photoreceptors (ON cells) or hyperpolarized when light hits photoreceptors (OFF cells). Bipolar cells communicate with RGCs by forming synapses with RGC dendrites in the inner plexiform layer (IPL). This layer is stratified primarily into two sections, an outer layer and an inner layer, where RGCs synapse with OFF and ON bipolar cells, respectively. There are three main types of RGCs – those that only synapse with ON or OFF bipolar cells (monostratified RGCs), as well as RGCs that synapse with both ON and OFF bipolar cells (bistratified RGCs).
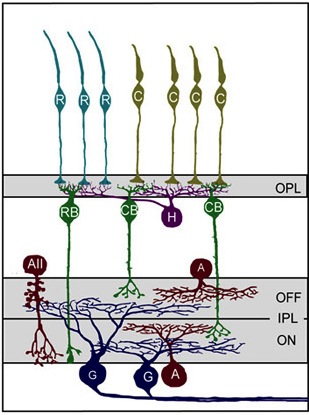
Figure 1: Model of primary cell types and their connection patterns in the mouse retina. First, Photoreceptors (teal rods (R) and yellowish cones, (C)) synapse with horizontal cells (purple) and bipolar cells (RB and CB, green) in the outer plexiform layer (OPL). Next bipolar cells synapse with ganglion cells (G, blue) in the inner plexiform layer (IPL). Amacrine cells (red) also synapse with both ganglion cells and bipolar cells in the IPL. Notice the stratification within the IPL based on the ON/OFF status of the bipolar cells. (Reproduced for educational purposes from Development of cell types and synaptic connections in the retina (http://webvision.med.utah.edu/).
During retinal development, RGCs form stratified dendritic arbors with synaptic connections to either exclusively ON bipolar cells (ON RGCs), exclusively OFF bipolar cells (OFF RGCs), or ON and OFF bipolar cells (ON-OFF RGCs). In the latter, RGC dendrites form synapses with ON and OFF bipolar cells such that ON and OFF inputs reside on separate laminar arborizations (e.g., Figure 1). Typically, ON and OFF bipolar cells form similar numbers of synapses with RGC dendrites, regardless of whether the RGCs are monostratified (ON or OFF) or bistratified (ON-OFF).
Neuronal Activity Determines Synapse Density within ON and OFF Layers of the IPL
To test whether this classical “use it or lose it” model for developing neuronal connections held true in the retina, Kerschensteiner et al. created a new transgenic line of mice that were unable to release glutamate from their ON bipolar cells, which is crucial for signaling to RGCs. The authors made a transgenic mouse that coupled the transcription of TeNT, a bacterial protease that inhibits vesicle fusion (which is necessary for presynaptic signaling), with the promoter for mGluR6, a glutamate receptor expressed in ON bipolar cells but not in OFF bipolar cells. Thus, ON cells could not communicate with the RGCs, while OFF cells could. If the classical model explains the development of connections between bipolar cells and RGCs, one would expect retinas in the transgenic mice to have no or very few RGC – ON cell synapses, and many RGC – OFF cell synapses.
As expected, the resulting mice exhibited normal receptive field properties from OFF-RGC connections and reduced or absent responses from ON-RGC ones. The dendritic branching and stratification patterns as well as overall connectivity appeared identical in transgenic and wild-type mice. Normal RGC development in the presence of significantly reduced ON bipolar cell activity indicates that partner selection and dendritic stratification of post-synaptic RGCs occur regardless of bipolar cell activity. These data provide evidence that RGCs are programmed to synapse to either ON bipolar cells, OFF bipolar cells, or both, regardless of presynaptic glutamate release.
By quantifying the density of individual postsynaptic sites in RGCs adjacent to ON bipolar cells in transgenic mice, the authors found that loss of glutamate release correlated with fewer mature synapses adjacent to ON bipolar cells (~50% fewer synapses in transgenic compared to wild-type retinas, Figure 2B). In addition, as might be predicted from the “use it or lose it” model, the density of synapses in bistratified RGCs connecting with the silenced ON bipolar cells was decreased relative to the density of synapses in the arbors connecting with active OFF bipolar cells (Figure 2C).
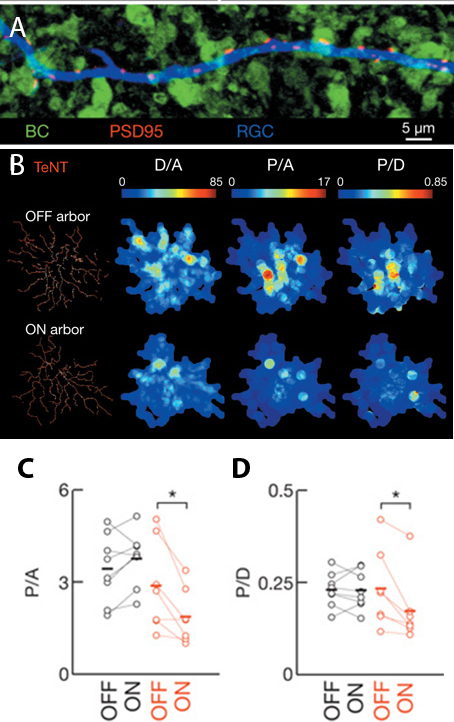
Figure 2: Density of bipolar-RGC synapses changes in response to glutamate release. A) Post-synaptic densities (PSD95, red) form at specific sites along RGC dendrites (blue) when closely apposed to biploar cell termini (green). B) Heat maps of glutamatergic post-synaptic densities for OFF and ON arborizations in bistratified RGCs. C-D) Graphical comparison of data in B. Values from non-transgenic retinas in black, transgenic animals in red. D/A is dendritic density, P/A is post-synaptic puncta per arbor area, P/D is post-synaptic puncta per dendritic length. Images are select panels from Figure 2 in Neurotransmission selectively regulates synapse formation in parallel circuits in vivo, Nature August 2009 (Vol 460, pp. 1016-1020). Kerschensteiner, D., Morgan, J., Parker, E., Lewis, R. & Wong, R. Used with permission of Nature Publishing Group.
When Kirchensteiner and coauthors examined the underlying mechanism for this decreased density, the “use it or lose it” model didn’t seem to hold true. Using time-lapse imaging, the authors showed that the silenced ON bipolar cells eliminated synapses at a rate comparable to wild-type but initial synapse formation was dramatically reduced. These data indicate that glutamate release is necessary for synapse formation and that elimination of synapses may not be directly tied to the activity of presynaptic input.
When the authors examined synapses at the ultrastructural level, they found even more evidence that “use it or lose it” isn’t the best analogy for synaptogenesis in the IPL. They show that ribbon synapses, the synaptic vesicle anchoring structures implicated in rapid neuronal transmission, are more numerous in the silent ON bipolar cells than in the active OFF bipolar cells (or in active ON bipolar cells in wild-type retinas). These intriguing electron micrographs raise the possibility that post-synaptic signaling, perhaps in response to glutamate, limits ribbon synapse formation.
With this study, Kerchensteiner et al. helped elucidate the role of glutamate release in forming synapses between bipolar cells and RGCs. Interestingly, glutamate release regulates how many synapses are formed between bipolar cells and RGCs but not how many synapses are eliminated, contrary to the classical “use it or lose it” model of synaptic formation. In a follow-up study, published in 2011 [5], some of the same authors investigate this teamwork approach to syanpatogensis in the IPL, providing evidence that differential synaptic maturation of axo-dendritic appositions is shaped by presynaptic activity and occurs in a cell type-specific manner. Whether (and how) post-synaptic cells may influence this process still seems to be an open question.
Although the “use it or lose it” analogy doesn’t completely describe what happens during synapse formation in the IPL, it does provide a framework for thinking about what cells need to connect with each other. In the case of bipolar cells and RGCs, it seems that glutamate needs to be used for efficient and robust synaptogenesis or the cells lose out all together, rarely making functional connections.
——————
[1]Kasthuri, N., & Lichtman, J. (2003). The role of neuronal identity in synaptic competition Nature, 424 (6947), 426-430 DOI: 10.1038/nature01836
[2]Wong, R. O. L. & Lichtman, J. W. in Fundamental Neuroscience 2nd edn (eds Squire, L. R. et al.) Ch. 20, 533–554 (Academic Press, 2002).
[3]MILLER, K. (1996). Synaptic Economics: Competition and Cooperation in Synaptic Plasticity Neuron, 17 (3), 371-374 DOI: 10.1016/S0896-6273(00)80169-5
[4] Kerschensteiner, D., Morgan, J., Parker, E., Lewis, R., & Wong, R. (2009). Neurotransmission selectively regulates synapse formation in parallel circuits in vivo Nature, 460 (7258), 1016-1020 DOI: 10.1038/nature08236
[5] Morgan, J., Soto, F., Wong, R., & Kerschensteiner, D. (2011). Development of Cell Type-Specific Connectivity Patterns of Converging Excitatory Axons in the Retina Neuron, 71 (6), 1014-1021 DOI: 10.1016/j.neuron.2011.08.025
 (1 votes)
(1 votes)
 Loading...
Loading...


 (1 votes)
(1 votes)

 The ability to reprogram human fibroblasts to neurons in vitro has opened up unprecedented opportunities in disease modelling and cellular therapeutics. Despite this breakthrough, a major challenge in the field is the limited phenotypic and functional maturation of the induced neurons (iNs). Now, on p.
The ability to reprogram human fibroblasts to neurons in vitro has opened up unprecedented opportunities in disease modelling and cellular therapeutics. Despite this breakthrough, a major challenge in the field is the limited phenotypic and functional maturation of the induced neurons (iNs). Now, on p. The intestinal stem cell (ISC) niche is responsible for coordinating the ongoing maintenance and regeneration of the adult gut. Wnt signalling is crucial for stem cell maintenance in the ISC niche, but the source of Wnts remains unclear. Now, on p.
The intestinal stem cell (ISC) niche is responsible for coordinating the ongoing maintenance and regeneration of the adult gut. Wnt signalling is crucial for stem cell maintenance in the ISC niche, but the source of Wnts remains unclear. Now, on p.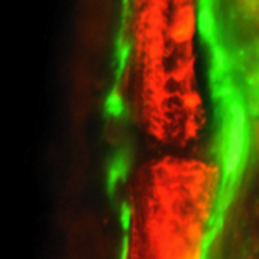 Complete bone regeneration following appendage amputation is remarkably efficient in zebrafish, but does not occur in mammals. Both groups, however, can repair bone fractures to varying extents, but whether a conserved cellular mechanism underpins both bone regeneration and repair in the adult zebrafish remains unclear. It is also unclear whether new tissue can be generated in other bony structures apart from the fin. In this issue (p.
Complete bone regeneration following appendage amputation is remarkably efficient in zebrafish, but does not occur in mammals. Both groups, however, can repair bone fractures to varying extents, but whether a conserved cellular mechanism underpins both bone regeneration and repair in the adult zebrafish remains unclear. It is also unclear whether new tissue can be generated in other bony structures apart from the fin. In this issue (p.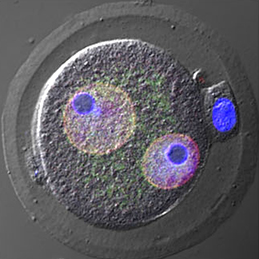 Almost every living cell contains a nucleus and, within that, a nucleolus. It is commonly accepted that the nucleolus of somatic cells arises from the nucleolar precursor body (NPB), a large, compact nucleolus present in the oocyte, but whether this is true remains to be definitively shown. Now, on p.
Almost every living cell contains a nucleus and, within that, a nucleolus. It is commonly accepted that the nucleolus of somatic cells arises from the nucleolar precursor body (NPB), a large, compact nucleolus present in the oocyte, but whether this is true remains to be definitively shown. Now, on p.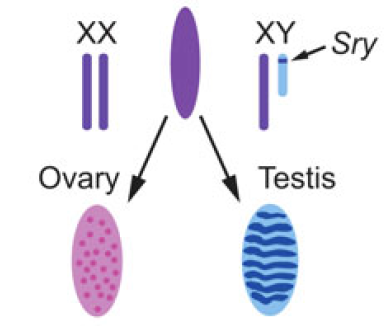 Christian Larney, Timothy Bailey and Peter Koopman review the transcriptional regulation of the testis-determining gene Sry. The authors present an integrated model for Sry regulation, and explain how this model functions to control sex determination in mammals. See the Review article on p.
Christian Larney, Timothy Bailey and Peter Koopman review the transcriptional regulation of the testis-determining gene Sry. The authors present an integrated model for Sry regulation, and explain how this model functions to control sex determination in mammals. See the Review article on p.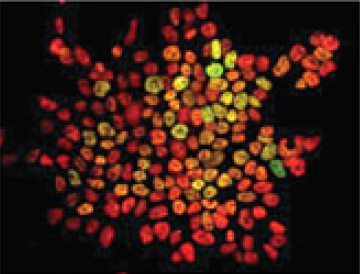 Maria-Elena Torres-Padilla and Ian Chambers review what is known about transcriptional heterogeneity in pluripotent stem cells, focusing on the underlying causes of heterogeneity and how transcriptional heterogeneity can be to the benefit of the whole stem cell population. See the Review article on p.
Maria-Elena Torres-Padilla and Ian Chambers review what is known about transcriptional heterogeneity in pluripotent stem cells, focusing on the underlying causes of heterogeneity and how transcriptional heterogeneity can be to the benefit of the whole stem cell population. See the Review article on p. (No Ratings Yet)
(No Ratings Yet)