I, Narender Kumar, am a graduate student in Prof John C. Larkin’s lab. Our lab is located in the Life Sciences Building on the main campus of Louisiana State University (LSU) Baton Rouge. LSU is located on the banks of the historical Mississippi River and the river levee is the one of the best places to walk or bike on a sunny winter’s day.
Arabidopsis thaliana (Fig.1A) is a model used to study plant genetics, molecular biology and biochemistry. This plant was first discovered by Johannes Thal in the sixteenth century and initially named “Pilosilla siliquosa”, but since then it has gone through a number of name changes and is now widely known as Arabidopsis thaliana, or by the common name thale cress or mouse-ear cress. The genus Arabidopsis has many species, but A. thaliana is the most commonly used in labs. This plant belongs to an agronomical important plant family- Brassicaceae (Cruciferae), though it does not have any agronomical importance in itself.
Arabidopsis trichome development is the main focus of our lab. The Arabidopsis trichome, or shoot epidermal hair, is a giant, unicellular polyploid cell on the leaf epidermis (Fig.1B). Trichomes are polyploid as a result of a process called endoreplication, a form of mitotic cell cycle in which DNA replicates without cytokinesis causing the cell to become polyploid. Our lab is trying to understand the mechanism of endoreplication using trichome development as a model.
Over the last 30 years, Arabidopsis use as a model has increased dramatically because of the following suitable characteristics: –
1) It has a short life cycle- Arabidopsis completes its life cycle in 8-12 weeks from germination to harvesting.
2) It easily grows in a restricted space and is very easy to maintain in an indoor growth chamber.
3) It produces many seeds. Each silique (seed capsule) contains 30-60 seeds, and each plant has around 50-60 siliques, so a plant can produce thousands of seeds.
4) It has a sequenced and comparatively small genome in the plant kingdom (135 megabases, and approximately 25000* genes). It has 5 chromosomes and has genes similar to those of agronomically important crops, so it is a good model for crop plants.
5) Transgenic lines are easily produced by infection with Agrobacterium tumefaciens (and the floral dip method used is easy and fast).
6) Numerous mutants are available in the stock centers: Arabidopsis Biological Resource Center (ABRC), Nottingham Arabidopsis Stock Center (NASC), and RIKEN Bioresource Center (BRC)/SENDAI Arabidopsis Seed Stock Center (SASSC) etc.
7) Arabidopsis is a self-pollinated plant and cross-pollination is easy to do in the lab.
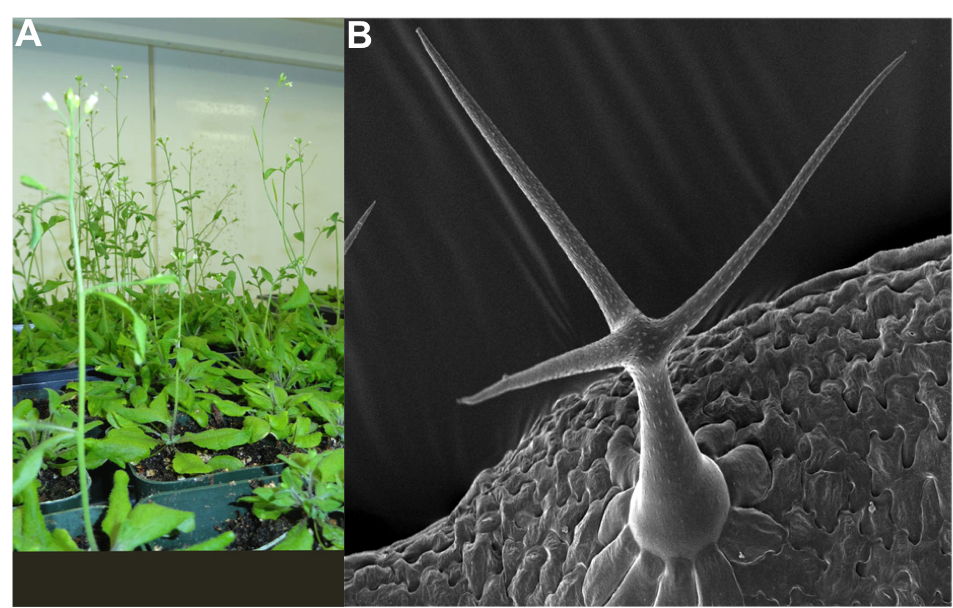
Fig-1. A) Arabidopsis plant in Growth chamber. B) Scanning Electron Microscopic Image (SEM) of Arabidopsis trichome
I am writing under the title “a day in the life of an Arabidopsis lab”, but it is very difficult to explain everything by just telling about one day. Therefore, to make it more understandable, I will explain step by step how I maintain and work with Arabidopsis throughout its life cycle.
Growth Chamber: – We have a very nice habitat (a climate-controlled chamber, Fig-2) for our favorite Arabidopsis plants in the basement of the Life Sciences Building. This chamber always maintains a 21-22 °C temperature and is divided into different shelves to house the different groups of plants. Besides the growth chamber, we have a preparation room in which we do all Arabidopsis related chores including sowing the seeds, harvesting and washing etc.

Fig-2. Climate-controlled growth chamber
Sowing the seeds: – In our lab, we grow Arabidopsis plants in big rectangular flats. Each flat can accommodate three black trays, and as each tray can hold 12 pots and each pot can have up to three plants, one flat can accommodate 36-108 plants (Fig-3A). However, when looking at seedling phenotypes, we plant approximately 30 seeds in each pot. The Arabidopsis life cycle starts with sowing the seeds on the soil. We fill the pots with ready-made dirt, water, and vermiculite to keep the soil well aerated.

Fig-3. A) A flat with 36 pots. B) MS media plates showing five days old Arabidopsis plants
Soil works best when growing transgenic plants that need to be sprayed (see later), but if I have any other marker (such as kanamycin) or need a clean root without soil or dirt to study root development then MS medium plates (Fig.3A) are ideal. Arabidopsis in MS plate plants are transferred to soil later. Arabidopsis seeds are very tiny and can easily be scattered around and contaminate other seeds.
Arabidopsis seeds need high humidity to germinate efficiently so the flats are covered with humidity domes until seeds germinate and start turning green, which takes five to six days (Fig-4A). I check the plants and growth chamber twice a day, in the morning and in the evening before I leave the lab. Plants must be watered properly as both too much water or too little water both can adversely affect plant growth. Too much water can lead to the appearance of white mold on the soil, which can infect the plants. Too little water can lead to drought stress. Therefore, proper watering and optimum temperature is necessary for a happily growing Arabidopsis plant. It is also important to protect the plants from insects, so we hang pest-traps in the growth chamber to catch fungus gnats.

Fig-4. A) Five-six old day plants. B) 15-day old plants. C) 1 month old plants
Flowering: – At approximately four-five weeks, plants start flowering and are ready for transformation to produce transgenic lines (Fig-4C). This means it is time to switch them from top to mid shelves. Only healthy looking flowering plants are used for transformation.
Transformation and separation of lines: – Tranformation is achieved by infecting seeds with Agrobacterium, which transfers the DNA of interest to the the developing ovule and produces transgenic seeds. Arabidopsis seeds are contained within siliques (seed capsules), so before every transformation I make sure to cut all the already-formed siliques to increase the efficiency of getting transgenic seeds. After transformation, I lay the plants down in a flat (Fig-5A) for two days, after which they are placed upright and watered with nutrient solution. One week after transformation the plants need to be stacked and tied (Fig-5B), both to support them and to avoid any contact or entanglement with other plants. Contamination is the main issue while maintaining a transgenic line, so I use Aracons (Fig-5C), which provide proper air to the plants and help avoiding the mixing of seeds to other lines.
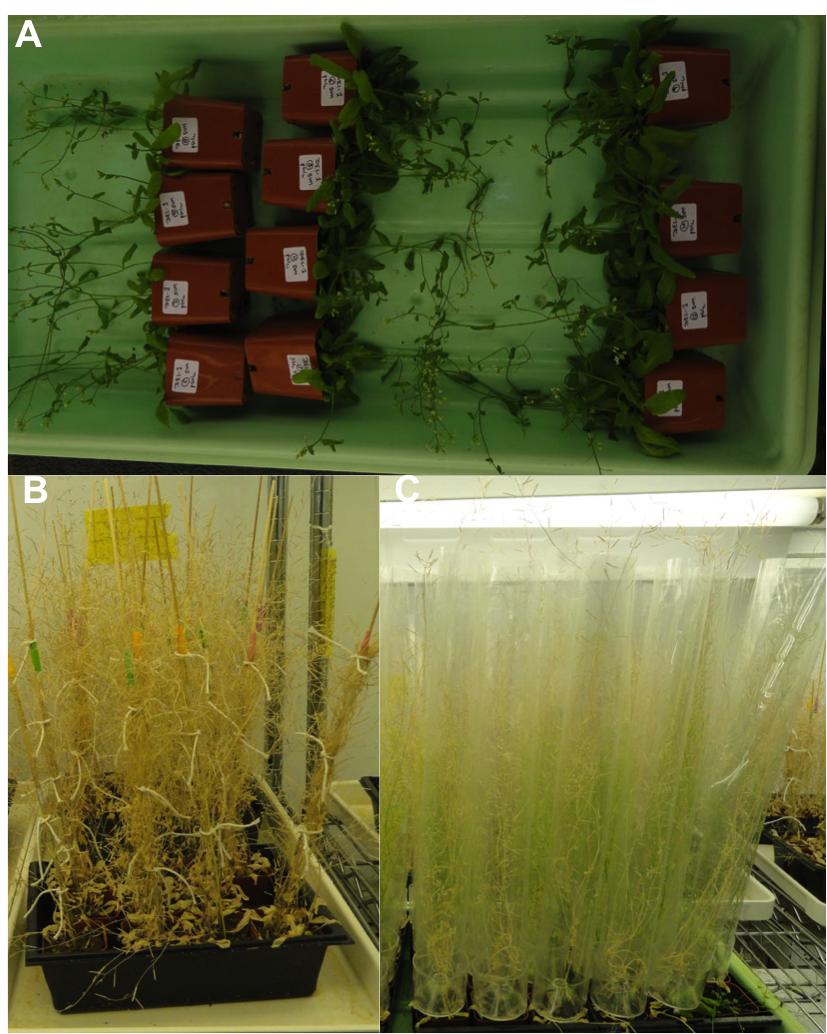
Fig-5. A) Plant after transformation. B) Tied plants after transformation. C) Plants in Aracons
Harvesting and store seeds: – After 3 or 4 weeks of transformation, plants become brown and dried, indicating that they are ready for harvesting (Fig-5B). Using scissors I carefully cut the plant from the pot, rub them with my fingers and strain the seeds to remove pods and plant debris (Fig-6). One plant can give thousands of seeds.

Fig-6. Harvesting tools
I always collect seeds in tubes containing 1 or 2 desiccant pellets. For long-term use, we store seeds in a desiccator at -20c. If seeds are not properly dry, over time their germination efficiency decreases.
Transgenic plants: – After collecting the transgenic seeds I and sow them in a big black tray (not in pots). Transgenic plants are resistant to the herbicide glufosinate-ammonium, so I spray the plants with this herbicide to select for successful transformants.
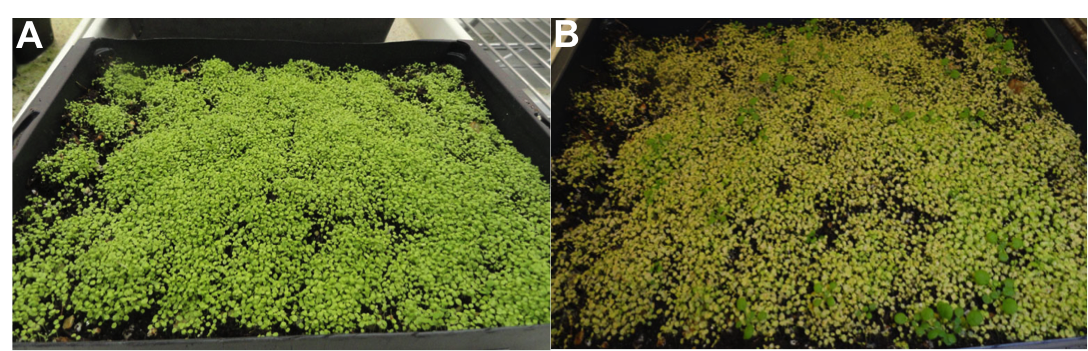
Fig-7. A) Seeds are planted for selection. B) Transgenic plants after selection
Advantages and disadvantages: – There is no need to be present around the clock, but the growth chamber has to be monitored frequently. There is no fear of any infection, blood or need for surgery, and it is a very clean and friendly model organism. With proper care, seeds will remain viable for years, so it is possible to return to old projects easily. The only major disadvantage is the chance of cross-contamination between seed lines. One line can be contaminated easily when other groups or two researchers are working on different project in the same area. Also, the generation time of six to eight weeks is a bit slower than other systems. But in the meantime, there are always molecular experiments to do.
*The number of genes in the Arabidopsis genome was edited from 2500 to 25000 (17/02/14)
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
 (14 votes)
(14 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)
 (3 votes)
(3 votes)






 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series 


 I think that one of the scariest parts of being President of the Society for Developmental Biology is coming up with topics for these editorials in the Newsletter.
I think that one of the scariest parts of being President of the Society for Developmental Biology is coming up with topics for these editorials in the Newsletter. (25 votes)
(25 votes)