What would happen to your memories and personality if, after decades of adult life, some portion of your brain was replaced with the progeny of fresh stem cells (as might happen in a treatment for degenerative brain disease)? Given the fascinating but poorly-understood examples of memory in aneural systems such as plants, ciliates, etc. (Eisenstein, 1975; Brugger et al., 2002; Volkov et al., 2008), is it possible that memories can be stored in tissues other than the brain? For that matter, how can arbitrary mental content (specific memories) be encoded and decoded within a physical medium such as a brain? What would be the dynamics of memories during brain regeneration?
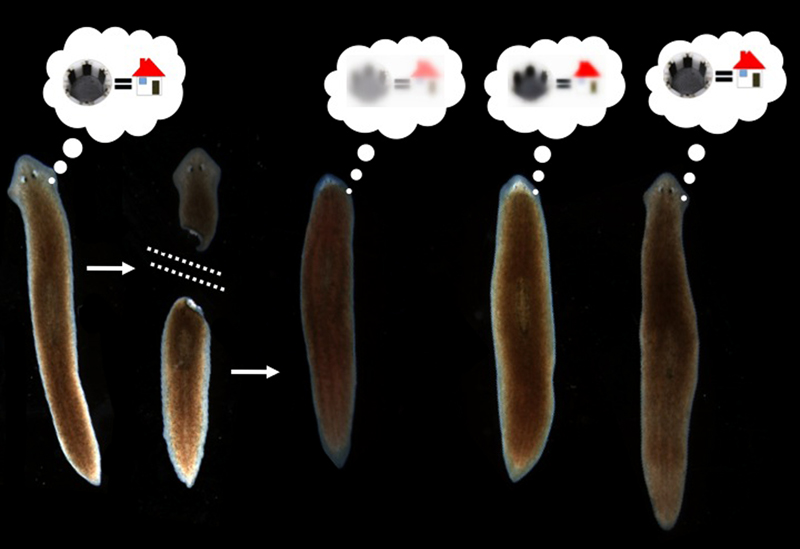
It has been shown that memories survive the drastic reorganization of the nervous system during metamorphosis from larva to adult in insects (Sheiman and Tiras, 1996; Blackiston et al., 2008). However, there is only one model system in which true memory and complete brain regeneration can be studied in the same animal: planaria. Planaria are free-living flatworms with bilateral symmetry, a true centralized brain, and a very rich behavioral repertoire. They are thus a popular system for the study of addiction and withdrawal. They also have remarkable powers of regeneration: a resident population of adult stem cells (neoblasts) enable these worms to regenerate any bodypart after surgical removal (Gentile et al., 2011; Lobo et al., 2012). In the 1960’s, a visionary named James V. McConnell experimentally posed a bizarre question (McConnell, 1965): if planaria were trained to form a specific memory and then regenerated their entire brain after decapitation, would the memories still be accessible?
McConnell’s group performed a number of experiments that suggested that the answer was Yes (McConnell et al., 1959; McConnell, 1966) – animals regenerating their whole head seemed to show recall of memories formed via several different training paradigms and behavioral assays. Unfortunately, at the time, training and testing experiments had to be done by hand. Manual experiments with worms are very tedious – not only time-consuming (resulting in low N’s and weak memories in worms who can only be trained for a short time per day) but also open to subjectivity during scoring of behaviors and very hard to control (behavioral experiments are notoriously sensitive to the skill of the operator). These difficulties led to the results being confirmed in some labs but not reproduced in some others (Corning, 1967; Corning and Riccio, 1970; Travis, 1981), and the whole area was largely forgotten (Rilling, 1996; Smalheiser et al., 2001) as people moved on to genetically-tractable model systems and easier questions of neuroscience.
In our lab, we are interested in information processing in cells and tissues. We study how cells orchestrate their activity towards maintaining and repairing complex anatomies, and how behavioral programs interact with radically-altered bodyplans (Levin, 2011). Indeed, our work has shown that all cells, not just excitable nerve, can communicate via changes of resting potential during patterning in embryogenesis, regeneration, and cancer suppression (Levin, 2012). This led to the hypothesis that perhaps networks of non-neural cells might also support the memory and information-processing abilities of neural nets; perhaps the ability of organisms to detect deviations from their target morphology and effect repairs is a reflection of true memory of specific shapes implemented by non-neural bioelectrical networks (Tseng and Levin, 2013)? Our work blurs the line between cognitive science and developmental biology, and we wondered if there may be mechanistic similarities between memory of spatial pattern (morphogenesis) in somatic structures, and memory of temporal patterns (learning and inference) in the brain. Aside from understanding development and developing interventions to increase regenerative ability, we are also interested in the synthetic bioengineering of hybrid tissues with new information-processing capabilities (developing new distributed computational platforms).
In order to establish a new model for studying encoding of information in living tissues and probe the mechanisms that interface the mind to the body, we turned to the planarian. Our first step was to build a next-generation testing and training device for flatworms (it also works for Xenopus and zebrafish (Blackiston and Levin, 2012; Blackiston and Levin, 2013)). The goal was to overcome prior roadblocks for this work: our system not only tracks the movement of planaria, but uniquely provides individual feedback (rewards and punishments) to 12 worms at a time, consistently (24*7 training), thus enabling not only behavior analysis but automated training in learning paradigms (Hicks et al., 2006; Blackiston et al., 2010). The idea was to do away with variability in manual training procedures, overcome operator tedium, and produce quantitative, objective, computer-scored analysis of worm performance after training. Five years and many engineering problems later, we had a device and a successful training protocol for planaria.
Amazingly, the data showed that if worms were trained to remember a novel kind of chamber environment with a specifically rough (laser-etched) surface, they would recognize it again (as measured by their willingness to eat a piece of liver rather than spend time exploring a new environment as controls do) after complete head removal and regeneration (Shomrat and Levin, 2013). While there is still plenty of room for improving the training protocol to induce an even more robust memory, and the worms need a brief refresher after head regeneration in order to show good recall of the original training, the results clearly showed that a complex brain-derived behavior was driven by a memory that survived complete head regeneration.
Our data suggest that not only can memories be stored somewhere outside of the brain, but that they can be imprinted on a naïve regenerated brain. Our future efforts will be focused on molecular cell biology and biophysics approaches to understand 1) which tissues contain the memory (e.g., are neoblasts required? Is it everywhere, or in certain regions?), 2) what molecular pathways underlie the imprinting of this information onto the brain, and 3) how specific memories are encoded into, and decoded from, living tissues. These data establish the automated analysis of memory in regenerated planaria as a new, highly tractable, model system in which to probe new aspects of cognitive science and the intersection of neurobiology and regeneration.
At the moment, we do not know where and how the memory is encoded in the body, or how prevalent such pathways are throughout the tree of life. However, based on exciting recent work on bioelectricity in somatic cells (McCaig et al., 2005; Allen et al., 2011; Bissiere et al., 2011; Wu et al., 2011; Tseng and Levin, 2013), we suggest the fascinating possibility that memory could be globally distributed throughout the body: non-neural cells communicating bioelectrically through gap junctions (electrical synapses) form a neural-like network that could be a very rich medium for encoding information and directing cell activity during regeneration (Chakravarthy and Ghosh, 1997; Bose and Karmakar, 2003; Inoue, 2008). Future work in this system will shed light not only on fundamental issues of memory but also on the biomedically-relevant questions of how regenerative therapies interact with cognitive content in patients. There will also likely be interesting applications of hybrid or biologically-inspired technologies for computational tissues and new computer architectures (Costello et al., 2011; Holley et al., 2011; Adamatzky, 2012; Adamatzky et al., 2012) that we can yet only imagine in their vaguest form.
References Cited
Adamatzky, A. (2012) ‘Slime mold solves maze in one pass, assisted by gradient of chemo-attractants’, IEEE transactions on nanobioscience 11(2): 131-4.
Adamatzky, A., Holley, J., Dittrich, P., Gorecki, J., De Lacy Costello, B., Zauner, K. P. and Bull, L. (2012) ‘On architectures of circuits implemented in simulated Belousov-Zhabotinsky droplets’, Biosystems 109(1): 72-7.
Allen, K., Fuchs, E. C., Jaschonek, H., Bannerman, D. M. and Monyer, H. (2011) ‘Gap Junctions between Interneurons Are Required for Normal Spatial Coding in the Hippocampus and Short-Term Spatial Memory’, J Neurosci 31(17): 6542-52.
Bissiere, S., Zelikowsky, M., Ponnusamy, R., Jacobs, N. S., Blair, H. T. and Fanselow, M. S. (2011) ‘Electrical synapses control hippocampal contributions to fear learning and memory’, Science 331(6013): 87-91.
Blackiston, D., Shomrat, T., Nicolas, C. L., Granata, C. and Levin, M. (2010) ‘A second-generation device for automated training and quantitative behavior analyses of molecularly-tractable model organisms’, PLoS ONE 5(12): e14370.
Blackiston, D. J. and Levin, M. (2012) ‘Aversive training methods in Xenopus laevis: general principles’, Cold Spring Harbor Protocols 2012(5).
Blackiston, D. J. and Levin, M. (2013) ‘Ectopic eyes outside the head in Xenopus tadpoles provide sensory data for light-mediated learning’, The Journal of experimental biology 216(Pt 6): 1031-40.
Blackiston, D. J., Silva Casey, E. and Weiss, M. R. (2008) ‘Retention of memory through metamorphosis: can a moth remember what it learned as a caterpillar?’, PLoS ONE 3(3): e1736.
Bose, I. and Karmakar, R. (2003) ‘Simple models of plant learning and memory’, Physica Scripta T106: 9-12.
Brugger, P., Macas, E. and Ihlemann, J. (2002) ‘Do sperm cells remember?’, Behav Brain Res 136(1): 325-8.
Chakravarthy, S. V. and Ghosh, J. (1997) ‘On Hebbian-like adaptation in heart muscle: a proposal for ‘cardiac memory”, Biol Cybern 76(3): 207-15.
Corning, W. C. (1967) ‘Regeneration and retention of acquired information’, NASA.
Corning, W. C. and Riccio, D. (1970) The planarian controversy, (ed. W. Byrne): New York: Academic Press.
Costello, B., Adamatzky, A., Jahan, I. and Zhang, L. A. (2011) ‘Towards constructing one-bit binary adder in excitable chemical medium’, Chemical Physics 381(1-3): 88-99.
Eisenstein, E. M. (1975) Aneural organisms in neurobiology, New York: Plenum Press.
Gentile, L., Cebria, F. and Bartscherer, K. (2011) ‘The planarian flatworm: an in vivo model for stem cell biology and nervous system regeneration’, Dis Model Mech 4(1): 12-9.
Hicks, C., Sorocco, D. and Levin, M. (2006) ‘Automated analysis of behavior: A computer-controlled system for drug screening and the investigation of learning’, J Neurobiol 66(9): 977-90.
Holley, J., Jahan, I., Costello Bde, L., Bull, L. and Adamatzky, A. (2011) ‘Logical and arithmetic circuits in Belousov-Zhabotinsky encapsulated disks’, Phys Rev E Stat Nonlin Soft Matter Phys 84(5 Pt 2): 056110.
Inoue, J. (2008) ‘A simple Hopfield-like cellular network model of plant intelligence’, Prog Brain Res 168: 169-74.
Levin, M. (2011) ‘The wisdom of the body: future techniques and approaches to morphogenetic fields in regenerative medicine, developmental biology and cancer’, Regenerative medicine 6(6): 667-73.
Levin, M. (2012) ‘Molecular bioelectricity in developmental biology: new tools and recent discoveries: control of cell behavior and pattern formation by transmembrane potential gradients’, Bioessays 34(3): 205-17.
Lobo, D., Beane, W. S. and Levin, M. (2012) ‘Modeling planarian regeneration: a primer for reverse-engineering the worm’, PLoS Comput Biol 8(4): e1002481.
McCaig, C. D., Rajnicek, A. M., Song, B. and Zhao, M. (2005) ‘Controlling cell behavior electrically: current views and future potential’, Physiol Rev 85(3): 943-78.
McConnell, J. V. (1965) A Manual of Psychological Experimentation on Planarians. Ann Arbor, Michigan: The Worm Runner’s Digest.
McConnell, J. V. (1966) ‘Comparative physiology: learning in invertebrates’, Annual Review of Physiology 28: 107-36.
McConnell, J. V., Jacobson, A. L. and Kimble, D. P. (1959) ‘The effects of regeneration upon retention of a conditioned response in the planarian’, Journal of Comparative Physiology and Psychology 52: 1-5.
Rilling, M. (1996) ‘The mystery of the vanished citations: James McConnell’s forgotten 1960s quest for planarian learning, a biochemical engram, and celebrity (vol 51, pg 589, 1996)’, American Psychologist 51(10): 1039-1039.
Sheiman, I. M. and Tiras, K. L. (1996) Memory and morphogenesis in planaria and beetle. in C. I. Abramson Z. P. Shuranova and Y. M. Burmistrov (eds.) Russian contributions to invertebrate behavior. Westport, CT: Praeger.
Shomrat, T. and Levin, M. (2013) ‘An automated training paradigm reveals long-term memory in planaria and its persistence through head regeneration’, J Exp Biol.
Smalheiser, N. R., Manev, H. and Costa, E. (2001) ‘RNAi and brain function: was McConnell on the right track?’, Trends Neurosci 24(4): 216-8.
Travis, G. D. L. (1981) ‘Aspects of the Social Construction of Learning in Planarian Worms’, Social Studies of Science 11: 11-32.
Tseng, A. and Levin, M. (2013) ‘Cracking the bioelectric code: Probing endogenous ionic controls of pattern formation’, Communicative & Integrative Biology 6(1): 1-8.
Volkov, A. G., Carrell, H., Adesina, T., Markin, V. S. and Jovanov, E. (2008) ‘Plant electrical memory’, Plant Signal Behav 3(7): 490-2.
Wu, C. L., Shih, M. F., Lai, J. S., Yang, H. T., Turner, G. C., Chen, L. and Chiang, A. S. (2011) ‘Heterotypic Gap Junctions between Two Neurons in the Drosophila Brain Are Critical for Memory’, Curr Biol.
Michael Levin,
Tufts University
www.drmichaellevin.org/
Tal Shomrat,
Hebrew University

 (5 votes)
(5 votes)
 Loading...
Loading...



 (1 votes)
(1 votes)






 Mutation of neurofibromatosis 2 (NF2) results in nervous system tumours. Molecularly, Nf2 has diverse functions, regulating cell-cell junction formation and various signalling pathways, including the Hippo-Yap pathway. However, the roles of Nf2 in the nervous system, and how its loss promotes tumorigenesis, are poorly understood. Here (
Mutation of neurofibromatosis 2 (NF2) results in nervous system tumours. Molecularly, Nf2 has diverse functions, regulating cell-cell junction formation and various signalling pathways, including the Hippo-Yap pathway. However, the roles of Nf2 in the nervous system, and how its loss promotes tumorigenesis, are poorly understood. Here ( During embryogenesis, transcriptional regulation must be coordinated with growth and cell division, so that genes are turned on or off in the right cells at the right time. Arjun Raj and colleagues now investigate the coupling of gene expression and cell division in C. elegans (
During embryogenesis, transcriptional regulation must be coordinated with growth and cell division, so that genes are turned on or off in the right cells at the right time. Arjun Raj and colleagues now investigate the coupling of gene expression and cell division in C. elegans ( Cells lining the lumen of various organs, such as the lung airway and the female reproductive tract, are multiciliated, and all the cilia are oriented in the same direction to generate flow. But how is cilia orientation coordinated within cells and across tissues? Chris Kintner and colleagues use the epithelial cells of Xenopus embryos as a model to study multicilate cell differentiation. On
Cells lining the lumen of various organs, such as the lung airway and the female reproductive tract, are multiciliated, and all the cilia are oriented in the same direction to generate flow. But how is cilia orientation coordinated within cells and across tissues? Chris Kintner and colleagues use the epithelial cells of Xenopus embryos as a model to study multicilate cell differentiation. On  Blood flow through the developing vasculature regulates vessel formation – both via the distribution of endocrine factors, and via mechanical force-induced responses. Several signalling pathways are known to be involved in this process, including signalling via the TGFb receptor Alk1, whose activity promotes quiescence in newly formed arteries and whose expression is itself dependent upon blood flow. On
Blood flow through the developing vasculature regulates vessel formation – both via the distribution of endocrine factors, and via mechanical force-induced responses. Several signalling pathways are known to be involved in this process, including signalling via the TGFb receptor Alk1, whose activity promotes quiescence in newly formed arteries and whose expression is itself dependent upon blood flow. On  Polycomb group proteins are chromatin regulators with highly conserved functions. The Polycomb repressive complex 2 (PRC2) methylates H3K27 to stably silence target genes, including the HOX genes in Drosophila. More recently, Utx and Jmjd3 demethylases were found to reverse PRC2-mediated H3K27 methylation, and it has been suggested that a dynamic cycle of methylation and demethylation is required for appropriate regulation of gene expression. Now, Ömer Copur and Jürg Müller challenge this view (
Polycomb group proteins are chromatin regulators with highly conserved functions. The Polycomb repressive complex 2 (PRC2) methylates H3K27 to stably silence target genes, including the HOX genes in Drosophila. More recently, Utx and Jmjd3 demethylases were found to reverse PRC2-mediated H3K27 methylation, and it has been suggested that a dynamic cycle of methylation and demethylation is required for appropriate regulation of gene expression. Now, Ömer Copur and Jürg Müller challenge this view (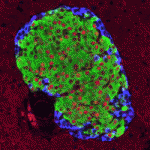 Integrins mediate cell-matrix adhesion and are also capable of inducing intracellular signalling cascades to regulate cell proliferation, differentiation and other cell behaviours. In vitro, disruption of β1 integrin function has been shown to affect various aspects of pancreatic β-cell activity. On
Integrins mediate cell-matrix adhesion and are also capable of inducing intracellular signalling cascades to regulate cell proliferation, differentiation and other cell behaviours. In vitro, disruption of β1 integrin function has been shown to affect various aspects of pancreatic β-cell activity. On 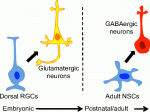 Eduardo Sequerro and colleagues review studies of postnatal and adult neurogenesis, challenging the notion that fixed genetic programs restrict neuronal fate. They hypothesize that the adult brain maintains plastic neural stem cells that are capable of responding to changes in environmental cues and generating diverse neuronal types. See the
Eduardo Sequerro and colleagues review studies of postnatal and adult neurogenesis, challenging the notion that fixed genetic programs restrict neuronal fate. They hypothesize that the adult brain maintains plastic neural stem cells that are capable of responding to changes in environmental cues and generating diverse neuronal types. See the 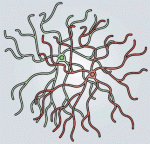 Weisheng Chen and Tom Maniatis provide a concise overview of the molecular and cellular biology of clustered Pcdhs, highlighting how they generate single cell diversity in the vertebrate nervous system and how such diversity may be used in neural circuit assembly. See the
Weisheng Chen and Tom Maniatis provide a concise overview of the molecular and cellular biology of clustered Pcdhs, highlighting how they generate single cell diversity in the vertebrate nervous system and how such diversity may be used in neural circuit assembly. See the  (No Ratings Yet)
(No Ratings Yet)