Amniote gastrulation without a streak
Posted by Cantas Alev, on 12 June 2013
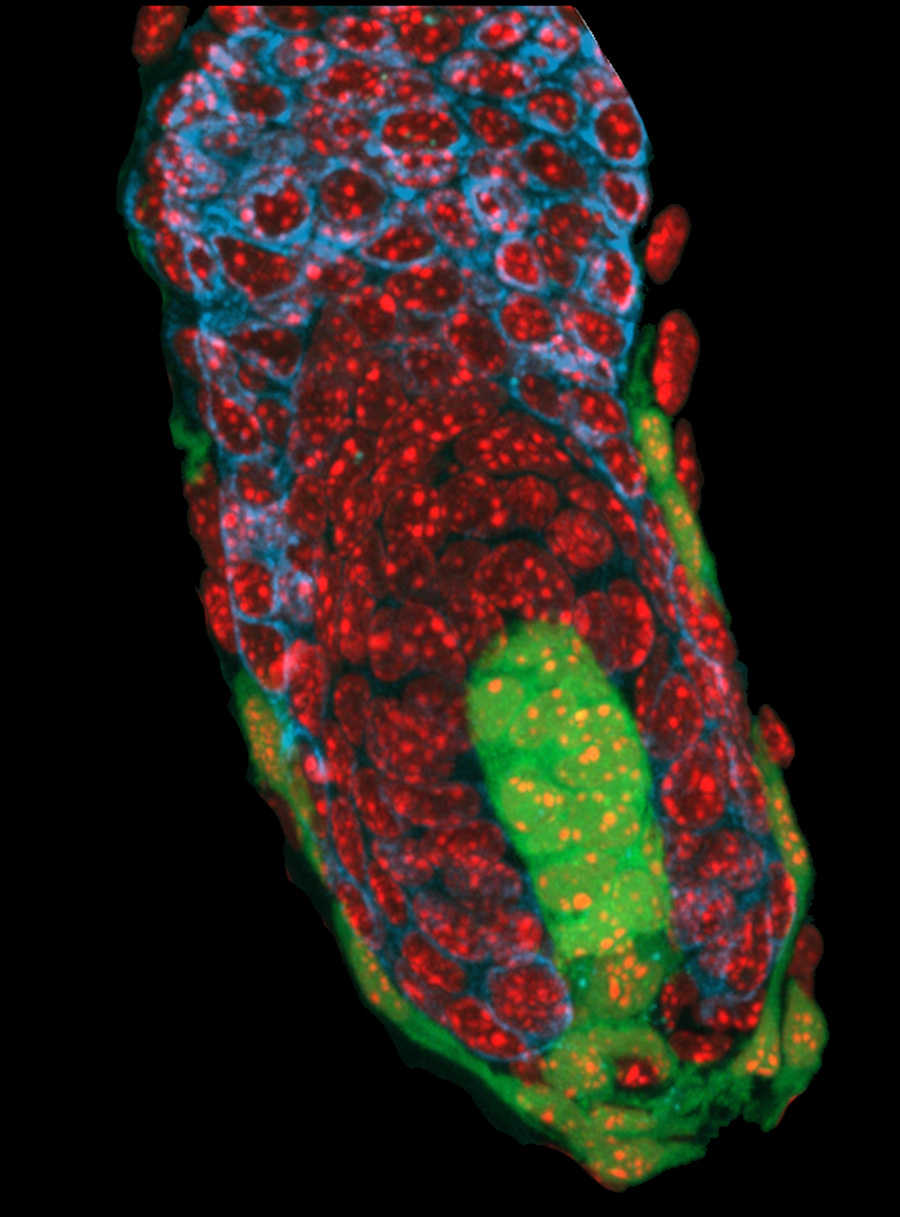
The three principal germ layers of the vertebrate embryo, ectoderm, mesoderm and endoderm, emerge from the pluripotent epiblast during the process of gastrulation. Being especially interested in the molecular and cellular mechanisms underlying the emergence of mesoderm, the source of a diverse set of tissues and cells including blood, muscle and bone, we decided to take a closer look at the primitive streak, the anatomical correlate of gastrulation in birds and mammals.
We initially set out to analyse the molecular mechanisms and signalling processes associated with formation and patterning of mesoderm in the primitive streak, reporting our findings in a previous Development paper (Alev et al., 2010). We then went on to tackle the long debated question about the evolutionary relationship between the primitive streak of amniotes and the blastopore of anamniotes (Shook and Keller, 2008), and asked whether it was possible to uncouple mesoderm induction and primitive streak formation. In order to efficiently address this question we needed a novel, simple and reproducible way to manipulate mesoderm differentiation and primitive streak formation, preferably in vivo, finally settling for in ovo subgerminal cavity injection. The subgerminal cavity is an ideal “reaction vessel” that can be used to assess the in vivo effects of growth factors and small molecules on the pre-gastrulation epiblast, as well as the differentiation of these cells giving rise to the germ-layers during gastrulation.
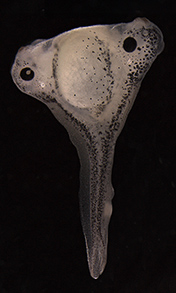
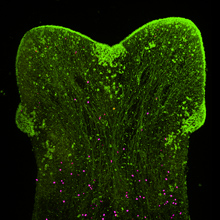
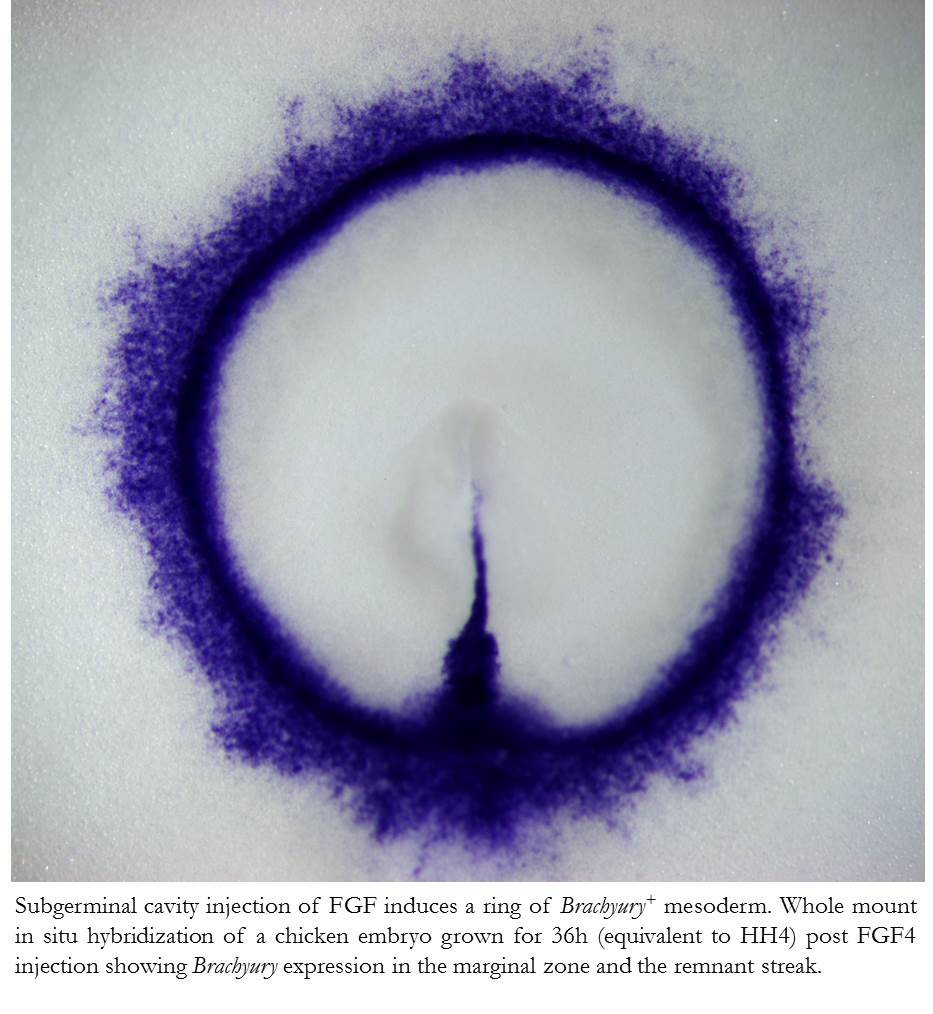
We found that subgerminal cavity injection of FGF can potently induce a ring of mesoderm in the marginal zone with more than half of the treated embryos having no primitive streak, which is generally assumed to be essential for gastrulation and mesoderm formation in birds. Further analysis of this unexpected “circumblastoporal” mode of mesoderm formation in chick revealed that the induced ring of mesoderm even had anamniote-type dorso-ventral polarity. Looking for an explanation as to why the induced mesoderm forms only in the outer margin of the epiblast (the marginal zone) while the center of the epiblast can not be turned into mesodermal precursors despite the uniform presence of FGF, we found that Wnt signalling, which is present in the marginal zone, is required not only for the circumblastoporal mode of mesoderm induction, but in concert with FGF can even turn the entire epiblast into mesodermal precursor fate. We also showed that TGFβ signalling contributes mainly to epithelial mesenchymal transition (EMT) and dorsalisation of the induced mesodermal precursors (Alev et al., 2013).
To further explore the possibilty that this hidden capacity for anamniote-type mesoderm formation might be also present in other amniotes, we tested the effects of subgerminal cavity injection of FGF in two other bird species: the quail, a close relative of chick; and emu, a basal ratite. We could induce circumblastoporal mesoderm in not only these bird species but could also generate a mesoderm ring in FGF-injected embryos of the Chinese soft-shelled turtle. Our observation that even a reptile, which does not possess a primitive streak to begin with, still maintains the ability for anamniote-type circumblastoporal mesoderm formation highlights the evolutionarily conserved nature of this phenomenon, supporting our hypothesis that the evolutionary emergence of amniotes was characterized by the restriction of a mesoderm inducing signal, likely FGF, to one side of the epiblast, while the overall capacity for anamniote-like circumblastoporal mesoderm formation was retained. Further support of our hypothesis may arise from studies of mesoderm induction in additional non-model organisms such as urodele and cacecilian amphibians and prototherian mammals.
Current model organisms are often selected out of experimental convenience rather than their necessarily being “model” representatives of the lineages they belong to, in contrast to the sheer variety of organisms studied during the height of comparative anatomy and embryology over a hundred years ago. It therefore doesn’t come as a surprise that even though a recent examination of reptilian gastrulation (Bertocchini et al., 2013) strongly suggests that the primitive streak is not a conserved feature among the amniotes, contrary statements still permeate vertebrate embryology textbooks. A revival of the utilisation of non-model organisms such as reptiles could thus help bypass the limitations of current models, especially in light of the recent advances made in the field of genomics. The usage of non-model-organisms in combination with comparative genomics and comparative embryology may thus lead to unexpected novel insights into questions old and new.
Alev, C., Wu, Y., Kasukawa, T., Jakt, L.M., Ueda, H.R., Sheng, G., 2010. Transcriptomic landscape of the primitive streak. Development 137, 2863-2874.
Alev, C., Wu, Y., Nakaya, Y., Sheng, G., 2013. Decoupling of amniote gastrulation and streak formation reveals a morphogenetic unity in vertebrate mesoderm induction. Development.
Bertocchini, F., Alev, C., Nakaya, Y., Sheng, G., 2013. A little winning streak: the reptilian-eye view of gastrulation in birds. Dev Growth Differ 55, 52-59.
Shook, D.R., Keller, R., 2008. Epithelial type, ingression, blastopore architecture and the evolution of chordate mesoderm morphogenesis. Journal of experimental zoology. Part B, Molecular and developmental evolution 310, 85-110.


 (8 votes)
(8 votes) (4 votes)
(4 votes)
 Asymmetric cell divisions (ACDs) play a crucial role in controlling cell fate and generating cell diversity during development. The centrosome is known to be involved in ACD, and recent studies have shown that centrosomes exhibit dynamic and asymmetric movements that regulate orientation of the mitotic spindle. Here, Yohanns Bellaiche and co-workers identify a novel type of centrosome movement during cytokinesis (
Asymmetric cell divisions (ACDs) play a crucial role in controlling cell fate and generating cell diversity during development. The centrosome is known to be involved in ACD, and recent studies have shown that centrosomes exhibit dynamic and asymmetric movements that regulate orientation of the mitotic spindle. Here, Yohanns Bellaiche and co-workers identify a novel type of centrosome movement during cytokinesis ( Early embryonic development occurs in the absence of transcription; instead, it relies on maternal mRNAs and proteins present within the egg. It is believed that this period of transcriptional quiescence is maintained by factors that eventually become titrated out during early cleavages, thus leading to zygotic genome activation. How exactly this transition occurs, however, is unclear. Here, Jim Smith, Steven Harvey and co-workers use exome sequencing and RNA-seq to distinguish between maternal and zygotic transcriptomes in early zebrafish embryos (
Early embryonic development occurs in the absence of transcription; instead, it relies on maternal mRNAs and proteins present within the egg. It is believed that this period of transcriptional quiescence is maintained by factors that eventually become titrated out during early cleavages, thus leading to zygotic genome activation. How exactly this transition occurs, however, is unclear. Here, Jim Smith, Steven Harvey and co-workers use exome sequencing and RNA-seq to distinguish between maternal and zygotic transcriptomes in early zebrafish embryos (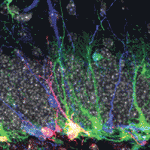 Neural stem/progenitor cells in the mammalian hippocampus generate new neurons throughout life. But how do these integrate into a mature and functional neural circuitry? Here, Sebastian Jessberger and colleagues address this question by using a new imaging approach to analyse neurite growth from newborn granular cells (
Neural stem/progenitor cells in the mammalian hippocampus generate new neurons throughout life. But how do these integrate into a mature and functional neural circuitry? Here, Sebastian Jessberger and colleagues address this question by using a new imaging approach to analyse neurite growth from newborn granular cells (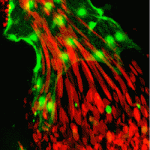 The correct formation of blood vessels is essential for the development of a functional vasculature. Various mechanisms of vascular lumen formation have been described to date but, now, Wiebke Herzog and colleagues examine the development of common cardinal veins (CCVs) in zebrafish and show that these form via a previously undescribed mode of lumen formation (
The correct formation of blood vessels is essential for the development of a functional vasculature. Various mechanisms of vascular lumen formation have been described to date but, now, Wiebke Herzog and colleagues examine the development of common cardinal veins (CCVs) in zebrafish and show that these form via a previously undescribed mode of lumen formation (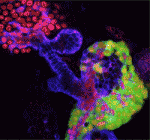 The liver and ventral pancreas are thought to develop from a common pool of multipotent progenitors. Although a number of studies have identified factors required for either pancreas or liver specification, factors that are distinctly required to specify the entire hepatopancreas system have not yet been reported. Now, Joseph Lancman and co-workers uncover a common genetic program, involving hnf1ba and wnt2bb, that specifies progenitors of the liver, ventral pancreas, gall bladder and associated ducts in zebrafish (
The liver and ventral pancreas are thought to develop from a common pool of multipotent progenitors. Although a number of studies have identified factors required for either pancreas or liver specification, factors that are distinctly required to specify the entire hepatopancreas system have not yet been reported. Now, Joseph Lancman and co-workers uncover a common genetic program, involving hnf1ba and wnt2bb, that specifies progenitors of the liver, ventral pancreas, gall bladder and associated ducts in zebrafish (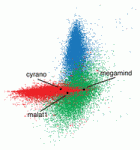 Long non-coding RNAs (lncRNAs) have recently emerged as key regulators of gene expression in embryos and in embryonic stem cells (ESCs). Recent large-scale genomics approaches have identified thousands of putative lncRNAs but are these all truly non-coding RNAs? Here, on
Long non-coding RNAs (lncRNAs) have recently emerged as key regulators of gene expression in embryos and in embryonic stem cells (ESCs). Recent large-scale genomics approaches have identified thousands of putative lncRNAs but are these all truly non-coding RNAs? Here, on  Song-Hai Shi and colleagues review recent findings on the generation, migration and organization of excitatory and inhibitory neurons in the neocortex, and discuss how the lineage history of neurons influences the assembly of functional circuits.
Song-Hai Shi and colleagues review recent findings on the generation, migration and organization of excitatory and inhibitory neurons in the neocortex, and discuss how the lineage history of neurons influences the assembly of functional circuits. Read the Editorial by our Editor-in-Chief, Olivier Pourquié on p.
Read the Editorial by our Editor-in-Chief, Olivier Pourquié on p. (No Ratings Yet)
(No Ratings Yet)