Here are the highlights from the new issue of Development:
Sniffing out sensory maps
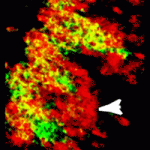 The development of sensory maps – neural representations of the sense organs – involves the growth of sensory nerve axons into specific regions of the brain and the formation of precise synaptic connections. On p. 2398, Jean-François Cloutier and colleagues investigate the molecular mechanism that underlies the coalescence of vomeronasal sensory neuron (VSN) axons into discrete synaptic units, termed glomeruli, in the mouse accessory olfactory bulb (AOB), which is involved in processing signals that control social and sexual interactions. The researchers report that the transmembrane proteins Kirrel2 and Kirrel3 are differentially expressed in subpopulations of VSNs and show that Kirrel3 expression is required for the coalescence of axons into glomeruli. Specifically, the posterior AOB in Kirrel3−/− mice contains fewer, larger glomeruli than the AOB of wild-type mice. Moreover, Kirrel3−/− mice display reduced male-male aggression. The researchers propose, therefore, that differential expression of Kirrels on VSN axons generates a molecular code that is required for the proper coalescence of these axons into glomeruli within the AOB.
The development of sensory maps – neural representations of the sense organs – involves the growth of sensory nerve axons into specific regions of the brain and the formation of precise synaptic connections. On p. 2398, Jean-François Cloutier and colleagues investigate the molecular mechanism that underlies the coalescence of vomeronasal sensory neuron (VSN) axons into discrete synaptic units, termed glomeruli, in the mouse accessory olfactory bulb (AOB), which is involved in processing signals that control social and sexual interactions. The researchers report that the transmembrane proteins Kirrel2 and Kirrel3 are differentially expressed in subpopulations of VSNs and show that Kirrel3 expression is required for the coalescence of axons into glomeruli. Specifically, the posterior AOB in Kirrel3−/− mice contains fewer, larger glomeruli than the AOB of wild-type mice. Moreover, Kirrel3−/− mice display reduced male-male aggression. The researchers propose, therefore, that differential expression of Kirrels on VSN axons generates a molecular code that is required for the proper coalescence of these axons into glomeruli within the AOB.
Tip-top role for SRF in angiogenesis
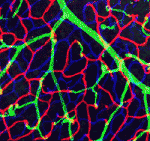 The evolutionarily conserved transcription factor SRF (serum response factor) is involved in several developmental processes that require cell migration, including embryonic angiogenesis. Here (p. 2321), Zhenlin Li and colleagues investigate the function of SRF during postnatal angiogenesis. They show that the inducible, endothelial-specific deletion of Srf in postnatal mice reduces postnatal growth and viability, induces systemic hypovascularisation and retinal angiopathy, and decreases angiogenesis in implanted tumours. Genetic mosaic analysis indicates that defective filopodia formation by the tip cells of angiogenic sprouts and reduced cell contractility are the primary causes of these angiogenic defects. The researchers also show that VEGFA induces nuclear accumulation of myocardin-related transcription factors (MRTFs; co-factors that regulate the transcriptional output of SRF) and that MRTF-SRF activity controls the expression of contractility genes that are important for endothelial cell migration. The researchers conclude that SRF regulates tip cell invasive behaviour during sprouting angiogenesis and hypothesize that curbing pathological angiogenesis by targeting the SRF pathway might restrict tumour growth.
The evolutionarily conserved transcription factor SRF (serum response factor) is involved in several developmental processes that require cell migration, including embryonic angiogenesis. Here (p. 2321), Zhenlin Li and colleagues investigate the function of SRF during postnatal angiogenesis. They show that the inducible, endothelial-specific deletion of Srf in postnatal mice reduces postnatal growth and viability, induces systemic hypovascularisation and retinal angiopathy, and decreases angiogenesis in implanted tumours. Genetic mosaic analysis indicates that defective filopodia formation by the tip cells of angiogenic sprouts and reduced cell contractility are the primary causes of these angiogenic defects. The researchers also show that VEGFA induces nuclear accumulation of myocardin-related transcription factors (MRTFs; co-factors that regulate the transcriptional output of SRF) and that MRTF-SRF activity controls the expression of contractility genes that are important for endothelial cell migration. The researchers conclude that SRF regulates tip cell invasive behaviour during sprouting angiogenesis and hypothesize that curbing pathological angiogenesis by targeting the SRF pathway might restrict tumour growth.
Pygopus, chromatin and Wnt signalling
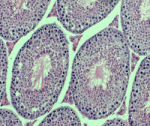 The mouse genome contains two genes for Pygopus (Pygo), which was identified as a Wnt signalling component in Drosophila. All Pygo proteins contain a conserved plant homology domain (PHD) that allows them to bind di- and trimethylated lysine 4 on histone H3, but is histone binding required for Pygo to function in Wnt signalling? To investigate this question, Konrad Basler and colleagues have generated mice homozygous for a Pygo2 mutation that abolishes chromatin binding (p. 2377). Surprisingly, Pygo2-chromatin binding is not needed to maintain Pygo2 function during mouse development, and abrogation of the Pygo2-chromatin interaction has little effect on Wnt signalling-related transcription during tissue homeostasis. Compromised Pygo2-chromatin binding leads to male sterility, however, and the researchers report that PHD-dependent recruitment of Pygo to regulatory regions in the testes is important for the recruitment of the histone acetylase Gcn5 to chromatin. These results identify a testis-specific role for Pygo2 as a chromatin remodeller that is unrelated to its role as a modulator of Wnt signalling.
The mouse genome contains two genes for Pygopus (Pygo), which was identified as a Wnt signalling component in Drosophila. All Pygo proteins contain a conserved plant homology domain (PHD) that allows them to bind di- and trimethylated lysine 4 on histone H3, but is histone binding required for Pygo to function in Wnt signalling? To investigate this question, Konrad Basler and colleagues have generated mice homozygous for a Pygo2 mutation that abolishes chromatin binding (p. 2377). Surprisingly, Pygo2-chromatin binding is not needed to maintain Pygo2 function during mouse development, and abrogation of the Pygo2-chromatin interaction has little effect on Wnt signalling-related transcription during tissue homeostasis. Compromised Pygo2-chromatin binding leads to male sterility, however, and the researchers report that PHD-dependent recruitment of Pygo to regulatory regions in the testes is important for the recruitment of the histone acetylase Gcn5 to chromatin. These results identify a testis-specific role for Pygo2 as a chromatin remodeller that is unrelated to its role as a modulator of Wnt signalling.
Tumours and embryos tread the same path
 Tumour cells share many characteristics with embryonic cells and it is thought that they acquire these characteristics through activation of developmental pathways. On p. 2354, Leonard Zon and co-workers develop a screening strategy to look for pathways that are common to embryogenesis and tumorigenesis in zebrafish. The researchers first evaluate gene expression in transgenic zebrafish embryos that express an inducible mutated RAS gene – RAS family members are the most commonly mutated oncogenes in human cancers and the RAS pathway is a key developmental pathway during embryogenesis. The researchers then use one of the genes that is upregulated by RAS expression to screen for small molecules that interfere with RAS signalling during embryogenesis. Finally, they show that two of the retrieved inhibitors suppress the growth of a zebrafish KRAS-induced embryonal rhabdomyosarcoma and of an NRAS-induced human rhabdomyosarcoma cell line. Together, these results demonstrate that common pathways are activated by RAS during embryogenesis and tumorigenesis and establish zebrafish embryos as a platform for anti-cancer drug discovery.
Tumour cells share many characteristics with embryonic cells and it is thought that they acquire these characteristics through activation of developmental pathways. On p. 2354, Leonard Zon and co-workers develop a screening strategy to look for pathways that are common to embryogenesis and tumorigenesis in zebrafish. The researchers first evaluate gene expression in transgenic zebrafish embryos that express an inducible mutated RAS gene – RAS family members are the most commonly mutated oncogenes in human cancers and the RAS pathway is a key developmental pathway during embryogenesis. The researchers then use one of the genes that is upregulated by RAS expression to screen for small molecules that interfere with RAS signalling during embryogenesis. Finally, they show that two of the retrieved inhibitors suppress the growth of a zebrafish KRAS-induced embryonal rhabdomyosarcoma and of an NRAS-induced human rhabdomyosarcoma cell line. Together, these results demonstrate that common pathways are activated by RAS during embryogenesis and tumorigenesis and establish zebrafish embryos as a platform for anti-cancer drug discovery.
Neural crest segregation out-Foxed
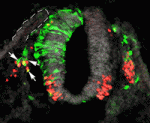 Fate segregation of multipotent progenitors is a key developmental process. The neural crest, which generates numerous cell types, is an ideal system in which to investigate this process. On p. 2269, Chaya Kalcheim and co-workers investigate when and how neurogenic and melanogenic neural crest cells segregate. Previously, the researchers reported that, even before emigration from the neural tube, neural but not melanocyte progenitors express the transcription factors Foxd3, Sox9 and Snail2. Now, they show that avian melanocytes are initially part of the Foxd3-positive premigratory epithelium but downregulate Foxd3 before emigration from the neural tube. If Foxd3 downregulation is prevented, avian melanocyte progenitors fail to upregulate the melanogenic marker Mitf and the guidance receptor Ednrb2. Consistent with these data, loss of Foxd3 function in mouse neural crest results in ectopic melanogenesis in the dorsal tube and sensory ganglia. The researchers propose, therefore, that Foxd3 is part of a dynamically expressed network of neural tube genes that regulates the segregation of neurogenic and melanogenic neural crest cells.
Fate segregation of multipotent progenitors is a key developmental process. The neural crest, which generates numerous cell types, is an ideal system in which to investigate this process. On p. 2269, Chaya Kalcheim and co-workers investigate when and how neurogenic and melanogenic neural crest cells segregate. Previously, the researchers reported that, even before emigration from the neural tube, neural but not melanocyte progenitors express the transcription factors Foxd3, Sox9 and Snail2. Now, they show that avian melanocytes are initially part of the Foxd3-positive premigratory epithelium but downregulate Foxd3 before emigration from the neural tube. If Foxd3 downregulation is prevented, avian melanocyte progenitors fail to upregulate the melanogenic marker Mitf and the guidance receptor Ednrb2. Consistent with these data, loss of Foxd3 function in mouse neural crest results in ectopic melanogenesis in the dorsal tube and sensory ganglia. The researchers propose, therefore, that Foxd3 is part of a dynamically expressed network of neural tube genes that regulates the segregation of neurogenic and melanogenic neural crest cells.
Elastic fibres form early in human cardiac valves
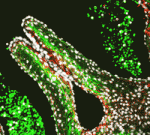 Cardiac semilunar valves, which stop blood leaking back into the heart, contain an elastic fibre network that is essential for their physiological function. Histochemical studies suggest that this network, which contains elastin, fibrillins, linking proteins such as fibulins, and fibronectin, is not crucial for early human cardiac valve development. Here (p. 2345), Katja Schenke-Layland and colleagues challenge this assumption by systematically analysing elastogenesis in human semilunar valves. The researchers report that the transcription of genes essential for elastic fibre formation starts early within the first trimester of pregnancy. Using immunohistochemistry, they show that fibronectin, fibrillins and linking proteins are present at the onset of cardiac cushion formation (about week 4 of development) and that tropoelastin/elastin protein expression is first detectable in the valves at about week 7 of pregnancy, the stage of development when the embryo’s blood pressure and heartbeat increase. Together, these findings suggest that elastogenesis is important for the development of functional semilunar valves.
Cardiac semilunar valves, which stop blood leaking back into the heart, contain an elastic fibre network that is essential for their physiological function. Histochemical studies suggest that this network, which contains elastin, fibrillins, linking proteins such as fibulins, and fibronectin, is not crucial for early human cardiac valve development. Here (p. 2345), Katja Schenke-Layland and colleagues challenge this assumption by systematically analysing elastogenesis in human semilunar valves. The researchers report that the transcription of genes essential for elastic fibre formation starts early within the first trimester of pregnancy. Using immunohistochemistry, they show that fibronectin, fibrillins and linking proteins are present at the onset of cardiac cushion formation (about week 4 of development) and that tropoelastin/elastin protein expression is first detectable in the valves at about week 7 of pregnancy, the stage of development when the embryo’s blood pressure and heartbeat increase. Together, these findings suggest that elastogenesis is important for the development of functional semilunar valves.
PLUS…
The neural crest
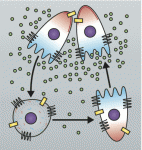 Roberto Mayor and Eric Theveneau provide an overview of neural crest formation, differentiation and migration, highlighting the molecular mechanisms governing neural crest migration.
Roberto Mayor and Eric Theveneau provide an overview of neural crest formation, differentiation and migration, highlighting the molecular mechanisms governing neural crest migration.
See the Development at a Glance poster article on p. 2247
Polar auxin transport: models and mechanisms
 Spatial patterns of auxin are important drivers of plant development. Here, ten Tusscher, Scheres and colleagues present a mathematical analysis of published models of auxin transport and suggest that current models alone cannot explain the auxin patterns found in plants. See the Hypothesis article on p. 2253
Spatial patterns of auxin are important drivers of plant development. Here, ten Tusscher, Scheres and colleagues present a mathematical analysis of published models of auxin transport and suggest that current models alone cannot explain the auxin patterns found in plants. See the Hypothesis article on p. 2253
 (1 votes)
(1 votes)
 Loading...
Loading...
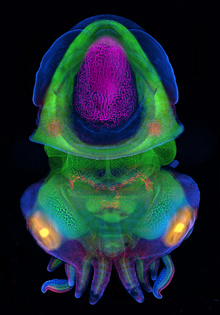
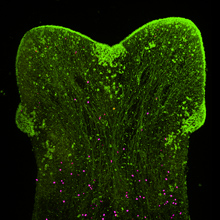
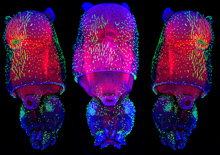
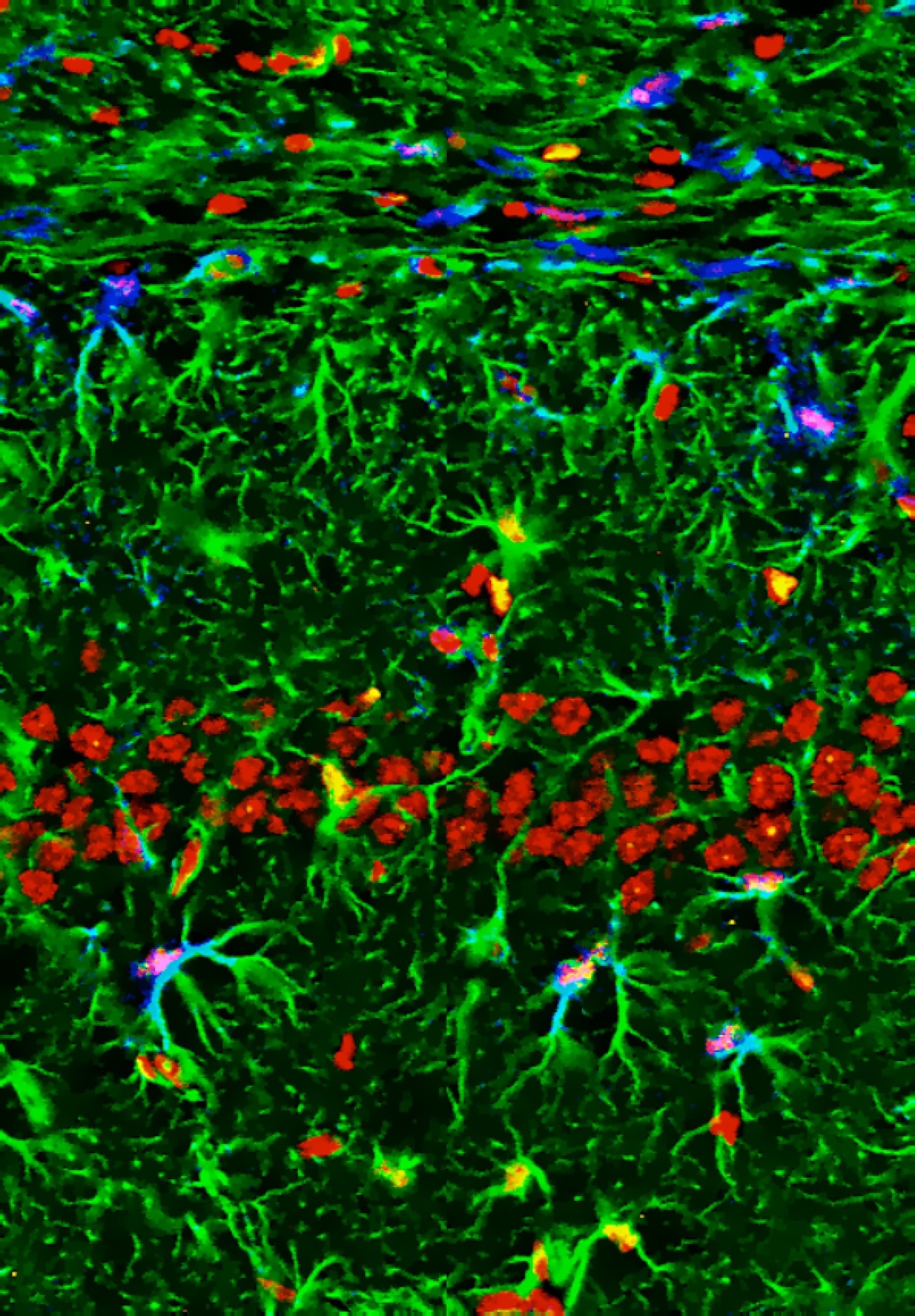
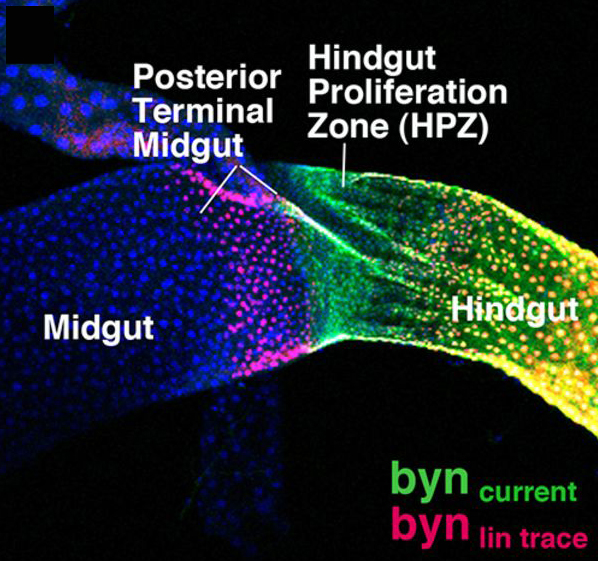 For most of us, we don’t all end up settled as adults in the same town where we were born. The same is true for many cells, including some stem cells in the fruit fly intestine. A recent paper describes the migration of progenitor cells, some of which will later become stem cells, in the intestine of developing flies, with the help of stunning images of these boundary-crossing cells.
For most of us, we don’t all end up settled as adults in the same town where we were born. The same is true for many cells, including some stem cells in the fruit fly intestine. A recent paper describes the migration of progenitor cells, some of which will later become stem cells, in the intestine of developing flies, with the help of stunning images of these boundary-crossing cells.![]() Takashima, S., Paul, M., Aghajanian, P., Younossi-Hartenstein, A., & Hartenstein, V. (2013). Migration of Drosophila intestinal stem cells across organ boundaries Development, 140 (9), 1903-1911 DOI: 10.1242/dev.082933
Takashima, S., Paul, M., Aghajanian, P., Younossi-Hartenstein, A., & Hartenstein, V. (2013). Migration of Drosophila intestinal stem cells across organ boundaries Development, 140 (9), 1903-1911 DOI: 10.1242/dev.082933

 (1 votes)
(1 votes)
 In other news from EuroStemCell, there’s lots of new material on our website:
In other news from EuroStemCell, there’s lots of new material on our website: (No Ratings Yet)
(No Ratings Yet)
 The development of sensory maps – neural representations of the sense organs – involves the growth of sensory nerve axons into specific regions of the brain and the formation of precise synaptic connections. On p.
The development of sensory maps – neural representations of the sense organs – involves the growth of sensory nerve axons into specific regions of the brain and the formation of precise synaptic connections. On p.  The evolutionarily conserved transcription factor SRF (serum response factor) is involved in several developmental processes that require cell migration, including embryonic angiogenesis. Here (p.
The evolutionarily conserved transcription factor SRF (serum response factor) is involved in several developmental processes that require cell migration, including embryonic angiogenesis. Here (p.  The mouse genome contains two genes for Pygopus (Pygo), which was identified as a Wnt signalling component in Drosophila. All Pygo proteins contain a conserved plant homology domain (PHD) that allows them to bind di- and trimethylated lysine 4 on histone H3, but is histone binding required for Pygo to function in Wnt signalling? To investigate this question, Konrad Basler and colleagues have generated mice homozygous for a Pygo2 mutation that abolishes chromatin binding (p.
The mouse genome contains two genes for Pygopus (Pygo), which was identified as a Wnt signalling component in Drosophila. All Pygo proteins contain a conserved plant homology domain (PHD) that allows them to bind di- and trimethylated lysine 4 on histone H3, but is histone binding required for Pygo to function in Wnt signalling? To investigate this question, Konrad Basler and colleagues have generated mice homozygous for a Pygo2 mutation that abolishes chromatin binding (p.  Tumour cells share many characteristics with embryonic cells and it is thought that they acquire these characteristics through activation of developmental pathways. On p.
Tumour cells share many characteristics with embryonic cells and it is thought that they acquire these characteristics through activation of developmental pathways. On p.  Fate segregation of multipotent progenitors is a key developmental process. The neural crest, which generates numerous cell types, is an ideal system in which to investigate this process. On p.
Fate segregation of multipotent progenitors is a key developmental process. The neural crest, which generates numerous cell types, is an ideal system in which to investigate this process. On p.  Cardiac semilunar valves, which stop blood leaking back into the heart, contain an elastic fibre network that is essential for their physiological function. Histochemical studies suggest that this network, which contains elastin, fibrillins, linking proteins such as fibulins, and fibronectin, is not crucial for early human cardiac valve development. Here (p.
Cardiac semilunar valves, which stop blood leaking back into the heart, contain an elastic fibre network that is essential for their physiological function. Histochemical studies suggest that this network, which contains elastin, fibrillins, linking proteins such as fibulins, and fibronectin, is not crucial for early human cardiac valve development. Here (p.  Roberto Mayor and Eric Theveneau provide an overview of neural crest formation, differentiation and migration, highlighting the molecular mechanisms governing neural crest migration.
Roberto Mayor and Eric Theveneau provide an overview of neural crest formation, differentiation and migration, highlighting the molecular mechanisms governing neural crest migration. Spatial patterns of auxin are important drivers of plant development. Here, ten Tusscher, Scheres and colleagues present a mathematical analysis of published models of auxin transport and suggest that current models alone cannot explain the auxin patterns found in plants. See the Hypothesis article on p.
Spatial patterns of auxin are important drivers of plant development. Here, ten Tusscher, Scheres and colleagues present a mathematical analysis of published models of auxin transport and suggest that current models alone cannot explain the auxin patterns found in plants. See the Hypothesis article on p. 
 – We had our first “
– We had our first “