Here are the highlights from the current issue of Development:
S1P1 halts vascular remodelling
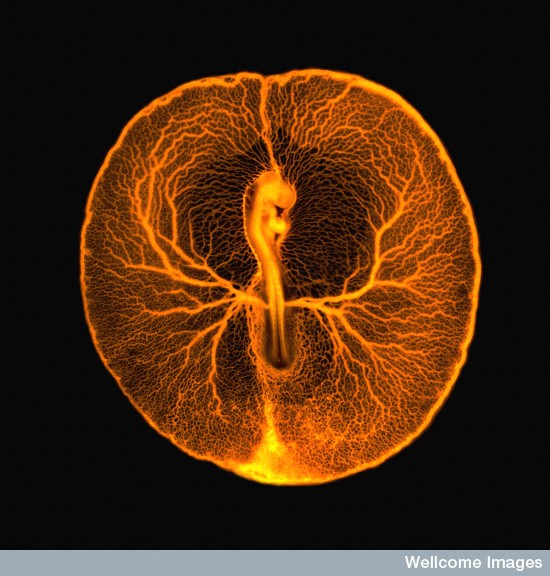
 During organogenesis, vascular remodelling ensures that developing organs receive sufficient oxygen and nutrients. Factors that induce sprouting angiogenesis, which is essential for vascular remodelling, have been identified, but the mechanisms that terminate sprouting angiogenesis, thereby stabilising the vasculature, remain unclear. Here (p. 3859), Adi Ben Shoham, Elazar Zelzer and colleagues report that sphingosine-1-phosphate receptor 1 (S1P1) inhibits sprouting angiogenesis during vascular development. S1P1 is known to mediate interactions between endothelial cells and mural cells during vascular maturation. Now, though, the researchers report that vessel size aberrations and excessive sprouting occur in the limbs of S1P1-null mice before vessel maturation, which suggests that S1P1 acts as an anti-angiogenic factor independently of mural cells. The effect of S1P1 on sprouting is endothelial cell-autonomous, they report, and similar vascular abnormalities develop in S1p1 knockdown zebrafish embryos. Because the S1P ligand is blood-borne, these findings suggest that blood flow completes a negative feedback loop that inhibits sprouting angiogenesis during development once the vascular bed is established and functional.
During organogenesis, vascular remodelling ensures that developing organs receive sufficient oxygen and nutrients. Factors that induce sprouting angiogenesis, which is essential for vascular remodelling, have been identified, but the mechanisms that terminate sprouting angiogenesis, thereby stabilising the vasculature, remain unclear. Here (p. 3859), Adi Ben Shoham, Elazar Zelzer and colleagues report that sphingosine-1-phosphate receptor 1 (S1P1) inhibits sprouting angiogenesis during vascular development. S1P1 is known to mediate interactions between endothelial cells and mural cells during vascular maturation. Now, though, the researchers report that vessel size aberrations and excessive sprouting occur in the limbs of S1P1-null mice before vessel maturation, which suggests that S1P1 acts as an anti-angiogenic factor independently of mural cells. The effect of S1P1 on sprouting is endothelial cell-autonomous, they report, and similar vascular abnormalities develop in S1p1 knockdown zebrafish embryos. Because the S1P ligand is blood-borne, these findings suggest that blood flow completes a negative feedback loop that inhibits sprouting angiogenesis during development once the vascular bed is established and functional.
Fruitful endoreduplication
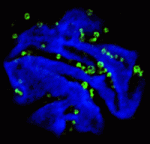 Endopolyploidy (increased cell ploidy) occurs during normal development in many eukaryotes. In higher plants, endopolyploidy is usually the result of endoreduplication – endonuclear DNA replication that produces chromosomes with multivalent chromatids. According to the ‘karyoplasmic ratio’ theory, a cell’s cytoplasmic volume is proportional to its nuclear DNA content. On p. 3817, Christian Chevalier and co-workers test this theory by analysing the structure of endoreduplicated nuclei in tomato fruit, which reach very high ploidy levels during their development. The researchers show that endopolyploidy in tomato pericarp (the fleshy part of the fruit) leads to the formation of polytene chromosomes. Pericarp nuclei, they report, have a complex structure in which numerous deep grooves are filled with mitochondria and in which there is a fairly constant ratio between nuclear surface area and the nuclear volume. Finally, they provide the first direct evidence that endoreduplication triggers enhanced transcription. Together, these results support the karyoplasmic theory and suggest that endoreduplication is associated with the complex cellular organisation that is required for tomato fruit development.
Endopolyploidy (increased cell ploidy) occurs during normal development in many eukaryotes. In higher plants, endopolyploidy is usually the result of endoreduplication – endonuclear DNA replication that produces chromosomes with multivalent chromatids. According to the ‘karyoplasmic ratio’ theory, a cell’s cytoplasmic volume is proportional to its nuclear DNA content. On p. 3817, Christian Chevalier and co-workers test this theory by analysing the structure of endoreduplicated nuclei in tomato fruit, which reach very high ploidy levels during their development. The researchers show that endopolyploidy in tomato pericarp (the fleshy part of the fruit) leads to the formation of polytene chromosomes. Pericarp nuclei, they report, have a complex structure in which numerous deep grooves are filled with mitochondria and in which there is a fairly constant ratio between nuclear surface area and the nuclear volume. Finally, they provide the first direct evidence that endoreduplication triggers enhanced transcription. Together, these results support the karyoplasmic theory and suggest that endoreduplication is associated with the complex cellular organisation that is required for tomato fruit development.
Cell-cell interactions set blastomere fate
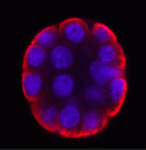 The inside-outside model of cell-fate specification in pre-implantation mammalian embryos proposes that blastomeres on the inside of the 16-cell stage embryo adopt an inner cell mass (ICM) fate, whereas those on the outside adopt a trophectoderm (TE) fate. Cell-cell contact should therefore be a key factor in this cell-fate specification event. On p. 3722, Chanchao Lorthongpanich and colleagues test this prediction by analysing the gene expression patterns of individual blastomeres separated from two-cell stage mouse embryos and re-separated after every subsequent cell division. Each singled blastomere has a unique gene expression pattern that is not characteristic of either ICM or TE, but that leans towards that of TE. Notably, embryos reconstructed from singled blastomeres are incapable of gastrulation but singled blastomeres preferentially contribute to the TE lineage when aggregated with intact embryos. Thus, the authors propose that a developmental clock drives the random expression of lineage-specific genes in pre-implantation embryos, but correct patterning of lineage-specific gene expression and proper embryonic development requires positional signals and cell-cell interactions.
The inside-outside model of cell-fate specification in pre-implantation mammalian embryos proposes that blastomeres on the inside of the 16-cell stage embryo adopt an inner cell mass (ICM) fate, whereas those on the outside adopt a trophectoderm (TE) fate. Cell-cell contact should therefore be a key factor in this cell-fate specification event. On p. 3722, Chanchao Lorthongpanich and colleagues test this prediction by analysing the gene expression patterns of individual blastomeres separated from two-cell stage mouse embryos and re-separated after every subsequent cell division. Each singled blastomere has a unique gene expression pattern that is not characteristic of either ICM or TE, but that leans towards that of TE. Notably, embryos reconstructed from singled blastomeres are incapable of gastrulation but singled blastomeres preferentially contribute to the TE lineage when aggregated with intact embryos. Thus, the authors propose that a developmental clock drives the random expression of lineage-specific genes in pre-implantation embryos, but correct patterning of lineage-specific gene expression and proper embryonic development requires positional signals and cell-cell interactions.
Epigenetic brain building
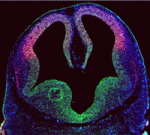 During brain development, neural progenitor cells (NPCs) give rise to various types of neurons and finally differentiate into astrocytes via switches in their differentiation competency. These switches involve changes in gene expression profiles that are thought to be governed partly by epigenetic control mechanisms, such as histone modification. Ryoichiro Kageyama and co-workers now report that the histone H3 Lys9 (H3K9) methyltransferase ESET (also known as Setdb1 or KMT1E) plays an essential role during mouse brain development (see p. 3806). ESET, they report, is highly expressed by mouse NPCs at early stages of brain development but is downregulated over time. Conditional ablation of ESET leads to reduced H3K9 trimethylation, misregulation of gene expression (including downregulation of neural gene expression, activation of non-neural gene expression and derepression of endogenous retroposons), severe brain defects and early lethality. Notably, loss of ESET impairs early neurogenesis but enhances astrocyte formation. Thus, the researchers suggest, ESET helps to regulate mouse brain development by epigenetically controlling temporal and tissue-specific gene expression.
During brain development, neural progenitor cells (NPCs) give rise to various types of neurons and finally differentiate into astrocytes via switches in their differentiation competency. These switches involve changes in gene expression profiles that are thought to be governed partly by epigenetic control mechanisms, such as histone modification. Ryoichiro Kageyama and co-workers now report that the histone H3 Lys9 (H3K9) methyltransferase ESET (also known as Setdb1 or KMT1E) plays an essential role during mouse brain development (see p. 3806). ESET, they report, is highly expressed by mouse NPCs at early stages of brain development but is downregulated over time. Conditional ablation of ESET leads to reduced H3K9 trimethylation, misregulation of gene expression (including downregulation of neural gene expression, activation of non-neural gene expression and derepression of endogenous retroposons), severe brain defects and early lethality. Notably, loss of ESET impairs early neurogenesis but enhances astrocyte formation. Thus, the researchers suggest, ESET helps to regulate mouse brain development by epigenetically controlling temporal and tissue-specific gene expression.
Replication origins wear many developmental HATs
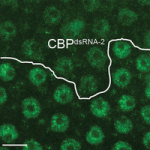 The genomic location and S-phase timing of origins of DNA replication change during multicellular development. Chromatin modifications influence differences in origin location and timing among different cells, but how is DNA replication coordinated with development programmes? Brian Calvi and colleagues have been examining developmental gene amplification in Drosophila ovarian follicle cells (p. 3880) and now report that the histone acetyltransferase (HAT) Chameau binds to amplicon origins and is partially required for their function. Unlike its human orthologue HBO1, however, Chameau is not absolutely required for gene amplification or genomic replication. The HAT CBP (Nejire) also binds to amplicon origins and is partially required for amplification, report the researchers, and Chameau and CBP collaborate in origin replication. Finally, Chameau and CBP globally regulate the developmental transition of follicle cells from endocycling to gene amplification. Thus, multiple HATs coordinate amplicon origin activity with follicle cell differentiation, and the researchers propose that origin regulation by multiple chromatin modifiers may be a general theme in development.
The genomic location and S-phase timing of origins of DNA replication change during multicellular development. Chromatin modifications influence differences in origin location and timing among different cells, but how is DNA replication coordinated with development programmes? Brian Calvi and colleagues have been examining developmental gene amplification in Drosophila ovarian follicle cells (p. 3880) and now report that the histone acetyltransferase (HAT) Chameau binds to amplicon origins and is partially required for their function. Unlike its human orthologue HBO1, however, Chameau is not absolutely required for gene amplification or genomic replication. The HAT CBP (Nejire) also binds to amplicon origins and is partially required for amplification, report the researchers, and Chameau and CBP collaborate in origin replication. Finally, Chameau and CBP globally regulate the developmental transition of follicle cells from endocycling to gene amplification. Thus, multiple HATs coordinate amplicon origin activity with follicle cell differentiation, and the researchers propose that origin regulation by multiple chromatin modifiers may be a general theme in development.
Developmentally, β-catenin levels matter
 β-Catenin is a central component of adherens junctions, which are required for cell sorting and migration during development, and of canonical Wnt signalling, which controls numerous developmental processes. Mice that lack β-catenin die before gastrulation but mice that express 50% of wild-type levels develop normally. Now, on p. 3711, Stefan Rudloff and Rolf Kemler examine the developmental consequences of other levels of β-catenin expression in mouse embryonic stem (ES) cells and embryos. Expression at ~12.5% of the wild-type level, they report, maintains the morphology and pluripotent characteristics of ES cells but cannot activate canonical target genes upon Wnt stimulation. Expression of β-catenin at ~25% of wild-type levels only partially restores Wnt signalling in vitro or in vivo. In embryos, they find that although both levels of expression partially rescue the knockout phenotype, neither results in proper gastrulation. Moreover, different Wnt targets require different β-catenin levels for expression at their wild-type levels. Thus, in mice, the level of β-catenin expression determines pluripotency, gastrulation and subsequent development.
β-Catenin is a central component of adherens junctions, which are required for cell sorting and migration during development, and of canonical Wnt signalling, which controls numerous developmental processes. Mice that lack β-catenin die before gastrulation but mice that express 50% of wild-type levels develop normally. Now, on p. 3711, Stefan Rudloff and Rolf Kemler examine the developmental consequences of other levels of β-catenin expression in mouse embryonic stem (ES) cells and embryos. Expression at ~12.5% of the wild-type level, they report, maintains the morphology and pluripotent characteristics of ES cells but cannot activate canonical target genes upon Wnt stimulation. Expression of β-catenin at ~25% of wild-type levels only partially restores Wnt signalling in vitro or in vivo. In embryos, they find that although both levels of expression partially rescue the knockout phenotype, neither results in proper gastrulation. Moreover, different Wnt targets require different β-catenin levels for expression at their wild-type levels. Thus, in mice, the level of β-catenin expression determines pluripotency, gastrulation and subsequent development.
Plus…
An excitingly predictable ‘omic future
Read the winning entry (written by Joanna Asprer) in the ‘Developments in development’ essay competition, which was run on the Node earlier this year.
Plant developmental biologists meet on stairways in Matera
 The third EMBO Conference on Plant Molecular Biology, which focused on ‘Plant development and environmental interactions’, was held in May 2012 in Matera, Italy. In this issue, Beeckman and Friml review some of the topics and themes that emerged from the meeting. See the Meeting Review on p. 3677
The third EMBO Conference on Plant Molecular Biology, which focused on ‘Plant development and environmental interactions’, was held in May 2012 in Matera, Italy. In this issue, Beeckman and Friml review some of the topics and themes that emerged from the meeting. See the Meeting Review on p. 3677
Stomatal development: a plant’s perspective on cell polarity, cell fate transitions and intercellular communication
 As part of our “Development: The Big Picture” series, Lau and Bergmann review how stomata can provide a conceptual and technical framework for the study of cell fate, stem cells, and cell polarity in plants. See the Primer on p. 3683
As part of our “Development: The Big Picture” series, Lau and Bergmann review how stomata can provide a conceptual and technical framework for the study of cell fate, stem cells, and cell polarity in plants. See the Primer on p. 3683
Ectodomain shedding and ADAMs in development
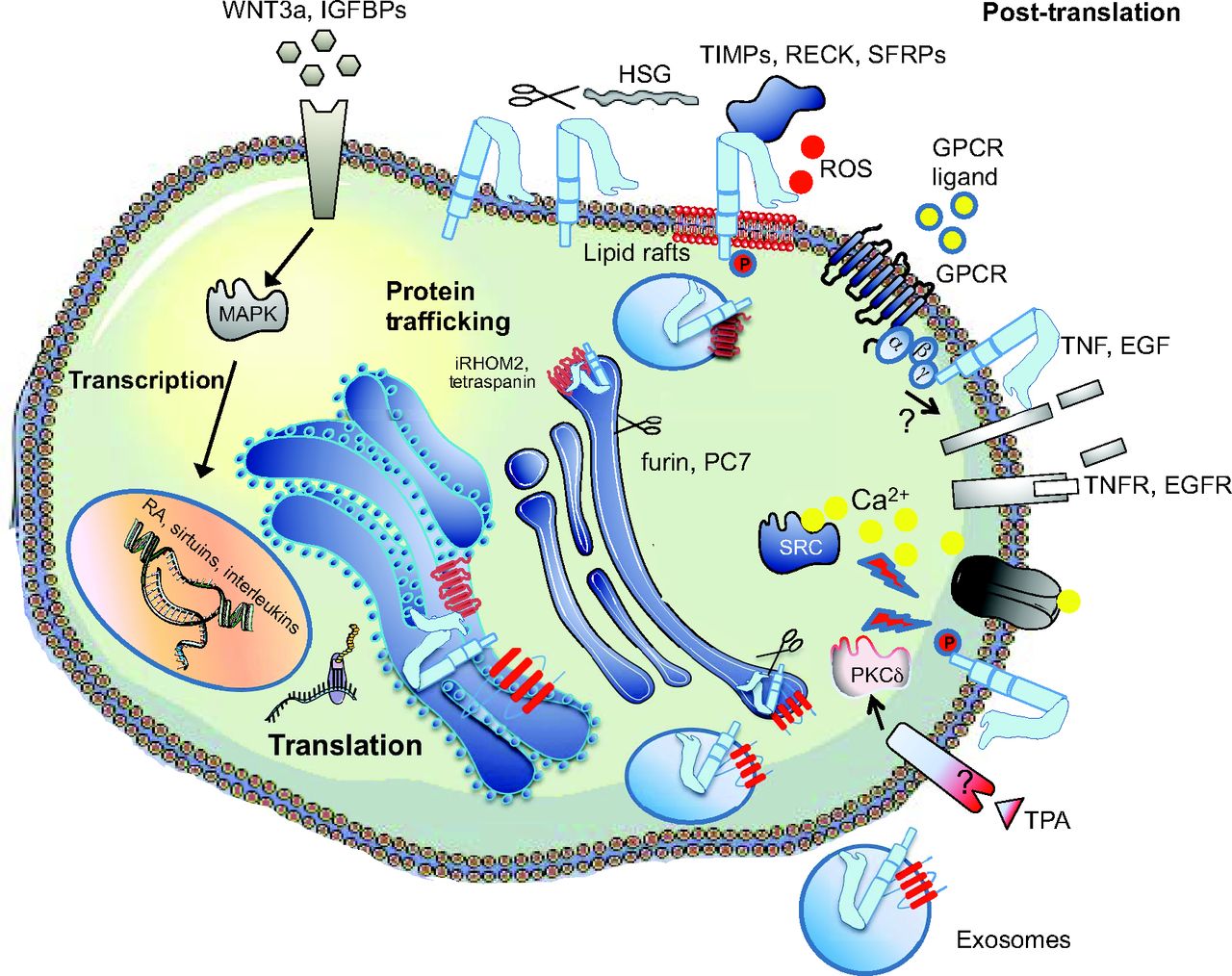 Weber and Saftig summarize the fascinating roles of A Disintegin And Metalloproteinases (ADAMs) in embryonic and adult tissue development in both vertebrates and invertebrates. See the Review on p. 3693
Weber and Saftig summarize the fascinating roles of A Disintegin And Metalloproteinases (ADAMs) in embryonic and adult tissue development in both vertebrates and invertebrates. See the Review on p. 3693
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)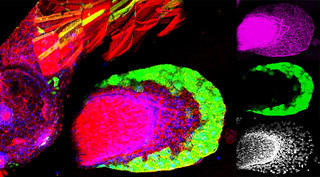



 (14 votes)
(14 votes) The fourth annual
The fourth annual  The evening closed with a reception and a visit to the exhibition area, where various companies and organisations have set up a stand for the next few days. There were many vendors and publishers, but also a few institutes: You can find out more about the future Francis Crick Centre in London, or about working at St Jude’s Children’s Hospital in Memphis. The Node is there, too, at the Company of Biologists booth just outside the exhibit hall (next to the job board), so do drop by if you’re attending. I’m there occasionally between talks and I love meeting Node readers.
The evening closed with a reception and a visit to the exhibition area, where various companies and organisations have set up a stand for the next few days. There were many vendors and publishers, but also a few institutes: You can find out more about the future Francis Crick Centre in London, or about working at St Jude’s Children’s Hospital in Memphis. The Node is there, too, at the Company of Biologists booth just outside the exhibit hall (next to the job board), so do drop by if you’re attending. I’m there occasionally between talks and I love meeting Node readers. (1 votes)
(1 votes) During organogenesis, vascular remodelling ensures that developing organs receive sufficient oxygen and nutrients. Factors that induce sprouting angiogenesis, which is essential for vascular remodelling, have been identified, but the mechanisms that terminate sprouting angiogenesis, thereby stabilising the vasculature, remain unclear. Here (p.
During organogenesis, vascular remodelling ensures that developing organs receive sufficient oxygen and nutrients. Factors that induce sprouting angiogenesis, which is essential for vascular remodelling, have been identified, but the mechanisms that terminate sprouting angiogenesis, thereby stabilising the vasculature, remain unclear. Here (p.  Endopolyploidy (increased cell ploidy) occurs during normal development in many eukaryotes. In higher plants, endopolyploidy is usually the result of endoreduplication – endonuclear DNA replication that produces chromosomes with multivalent chromatids. According to the ‘karyoplasmic ratio’ theory, a cell’s cytoplasmic volume is proportional to its nuclear DNA content. On p.
Endopolyploidy (increased cell ploidy) occurs during normal development in many eukaryotes. In higher plants, endopolyploidy is usually the result of endoreduplication – endonuclear DNA replication that produces chromosomes with multivalent chromatids. According to the ‘karyoplasmic ratio’ theory, a cell’s cytoplasmic volume is proportional to its nuclear DNA content. On p.  The inside-outside model of cell-fate specification in pre-implantation mammalian embryos proposes that blastomeres on the inside of the 16-cell stage embryo adopt an inner cell mass (ICM) fate, whereas those on the outside adopt a trophectoderm (TE) fate. Cell-cell contact should therefore be a key factor in this cell-fate specification event. On p.
The inside-outside model of cell-fate specification in pre-implantation mammalian embryos proposes that blastomeres on the inside of the 16-cell stage embryo adopt an inner cell mass (ICM) fate, whereas those on the outside adopt a trophectoderm (TE) fate. Cell-cell contact should therefore be a key factor in this cell-fate specification event. On p.  During brain development, neural progenitor cells (NPCs) give rise to various types of neurons and finally differentiate into astrocytes via switches in their differentiation competency. These switches involve changes in gene expression profiles that are thought to be governed partly by epigenetic control mechanisms, such as histone modification. Ryoichiro Kageyama and co-workers now report that the histone H3 Lys9 (H3K9) methyltransferase ESET (also known as Setdb1 or KMT1E) plays an essential role during mouse brain development (see p.
During brain development, neural progenitor cells (NPCs) give rise to various types of neurons and finally differentiate into astrocytes via switches in their differentiation competency. These switches involve changes in gene expression profiles that are thought to be governed partly by epigenetic control mechanisms, such as histone modification. Ryoichiro Kageyama and co-workers now report that the histone H3 Lys9 (H3K9) methyltransferase ESET (also known as Setdb1 or KMT1E) plays an essential role during mouse brain development (see p.  The genomic location and S-phase timing of origins of DNA replication change during multicellular development. Chromatin modifications influence differences in origin location and timing among different cells, but how is DNA replication coordinated with development programmes? Brian Calvi and colleagues have been examining developmental gene amplification in Drosophila ovarian follicle cells (p.
The genomic location and S-phase timing of origins of DNA replication change during multicellular development. Chromatin modifications influence differences in origin location and timing among different cells, but how is DNA replication coordinated with development programmes? Brian Calvi and colleagues have been examining developmental gene amplification in Drosophila ovarian follicle cells (p.  β-Catenin is a central component of adherens junctions, which are required for cell sorting and migration during development, and of canonical Wnt signalling, which controls numerous developmental processes. Mice that lack β-catenin die before gastrulation but mice that express 50% of wild-type levels develop normally. Now, on p.
β-Catenin is a central component of adherens junctions, which are required for cell sorting and migration during development, and of canonical Wnt signalling, which controls numerous developmental processes. Mice that lack β-catenin die before gastrulation but mice that express 50% of wild-type levels develop normally. Now, on p.  The third EMBO Conference on Plant Molecular Biology, which focused on ‘Plant development and environmental interactions’, was held in May 2012 in Matera, Italy. In this issue, Beeckman and Friml review some of the topics and themes that emerged from the meeting. See the Meeting Review on p.
The third EMBO Conference on Plant Molecular Biology, which focused on ‘Plant development and environmental interactions’, was held in May 2012 in Matera, Italy. In this issue, Beeckman and Friml review some of the topics and themes that emerged from the meeting. See the Meeting Review on p.  As part of our “
As part of our “ Weber and Saftig summarize the fascinating roles of A Disintegin And Metalloproteinases (ADAMs) in embryonic and adult tissue development in both vertebrates and invertebrates. See the Review on p.
Weber and Saftig summarize the fascinating roles of A Disintegin And Metalloproteinases (ADAMs) in embryonic and adult tissue development in both vertebrates and invertebrates. See the Review on p.