Here are the highlights from the current issue of Development:
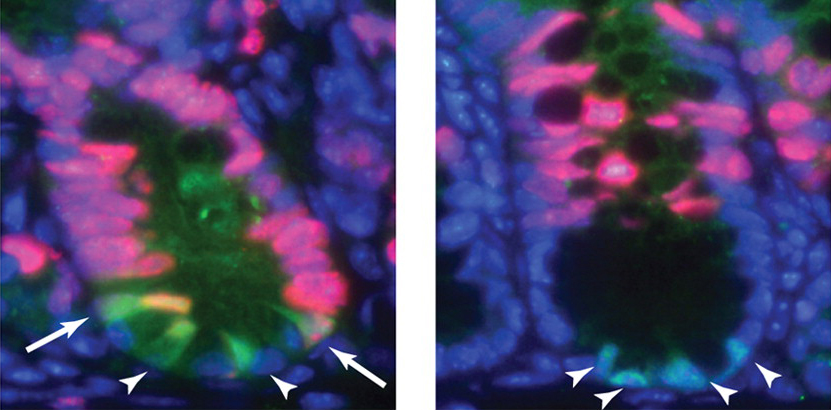
CycA takes control of endoreplication
Endocycles – repeated rounds of DNA replication without intervening mitoses – are involved in several terminal differentiation events. In Drosophila, for example, endoreplication occurs during the terminal differentiation of mechanosensory bristles. Endocycles are thought not to involve mitotic cyclins but here (p. 547), Agnès Audibert, Michel Gho and colleagues overturn that view by showing that cyclin A (CycA), which was thought to function exclusively in mitosis in Drosophila, is involved in endoreplication in the bristle lineage. The researchers show that CycA accumulates during the last part of endoreplication. CycA loss- and gain-of-function both induce changes in the dynamics of endoreplication, they report, and reduce the number of endocycles. Finally, CycA is required for relocalisation of ORC2, a member of the pre-replication complex, to the heterochromatin. These and other data reveal that CycA oscillations regulate endocycle dynamics in the fly mechanosensory bristle lineage and suggest that endoreplication involves remodelling of the entire cell-cycle network rather than simply a restriction of the canonical cell cycle as previously suggested.
Plane fact: aPKC orientates mitotic spindles
Mitotic spindle orientation, which is essential for epithelial morphogenesis and tissue maintenance, involves interactions between cortical polarity components and astral microtubules. The molecular machinery that regulates spindle apicobasal orientation during asymmetric cell division is well understood but what orientates the spindle along the epithelial plane in symmetrically dividing epithelial cells? On p. 503, Rui Gonçalo Martinho and colleagues provide the first in vivo evidence that atypical protein kinase C (aPKC) is involved in this process. Using a temperature-sensitive aPKC allele, the researchers show that Drosophila aPKC is required in imaginal discs for spindle planar orientation and for apical exclusion of Pins, a component of the molecular machinery that links the cell cortex to the astral microtubules. Apically localized aPKC is important for spindle planar orientation in mammalian epithelial cells in tissue culture; thus, these new observations suggest that the cortical cues required for spindle planar orientation are conserved between Drosophila and mammalian cells and are similar to those required for spindle orientation during asymmetric cell division.
pecanex wraps Notch up
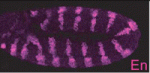
During early development of the Drosophila nervous system, Notch signalling limits neuroblast numbers by preventing the cells that neighbour neuroblasts from also choosing a neuroblast fate. Disruption of Notch signalling prevents this ‘lateral inhibition’ and produces hyperplasias of the embryonic nervous system. The absence of pecanex (pcx), which encodes a conserved multi-pass transmembrane protein of unknown function, also causes a similar neurogenic phenotype. Now, Kenji Matsuno and colleagues propose that Pcx is a novel component of the Notch signalling pathway in Drosophila (see p. 558). They show that Pcx resides in the endoplasmic reticulum (ER) and is required upstream of activated Notch. Disruption of pcx function, they report, results in ER enlargement. However, hyper-induction of the unfolded protein response in the absence of pcx suppresses both ER enlargement and the development of a neurogenic phenotype. Together, these results suggest that the ER plays a previously unrecognised role in Notch signalling that involves Notch folding and that this ER function depends on pcx activity.
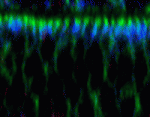
Wnt signalling regulates ciliogenesis

In zebrafish embryos, motile cilia lining the Kupffer’s vesicle (KV; the fish equivalent of the mouse node) help to establish left-right (LR) asymmetry. Wnt/β-catenin signalling is also involved in this process but precisely how it functions is unclear. Xueying Lin and colleagues now reveal that Wnt/β-catenin signalling directly regulates ciliogenesis in the zebrafish KV (see p. 514). The researchers show that reduced Wnt signalling disrupts LR patterning and ciliogenesis and downregulates Foxj1, a transcription factor that is required for the biosynthesis of motile cilia. KV-specific expression of foxj1a, they report, requires the presence of putative Lef1/Tcf binding sites in the foxj1a enhancer region, which suggests that Wnt signalling activates fox1ja transcription directly. Importantly, reduction of Wnt signalling also impairs foxj1 expression and ciliogenesis in developing zebrafish pronephric ducts and otic vesicles, epithelial structures that require Wnt activity for their development and function. The researchers propose, therefore, that the regulation of Foxj1 expression and ciliogenesis by Wnt/β-catenin signalling is a general developmental mechanism in zebrafish.
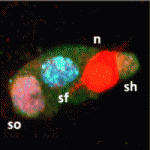
Egg cell orchestrates gametophyte development

Plant germ cells develop in specialised haploid structures called gametophytes. The female gametophyte of flowering plants contains an egg cell (which develops into the embryo), a central cell (which generates the endosperm that nurtures the embryo) and two accessory cell types, but what coordinates the development of these different cell types? On p. 498, Rita Groß-Hardt and co-workers report that egg-cell signalling mediated by LACHESIS (LIS), which encodes a homologue of a yeast pre-RNA splicing factor, regulates the development of Arabidopsis female gametophytic cells. lis mutants form supernumerary egg cells at the expense of accessory cells. The researchers now show that reducing LIS transcript levels specifically in the egg cell affects all the gametophyte cell types, which suggests that the egg cell orchestrates gametophyte differentiation. Notably, reduced LIS transcript levels in the egg cell interfere with homotypic nuclei fusion in the central cell and, consequently, endosperm formation. Thus, LIS-mediated egg-cell signalling ensures that endosperm only forms in the presence of a functional egg cell.
DAZL takes heat off testes
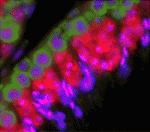
Mammalian testes are usually located outside the body cavity to ensure that the male germ cells are maintained at a low enough temperature to develop properly. If the testes get too warm, heat stress can cause germ cell apoptosis, which reduces fertility. Now, on p. 568, Kunsoo Rhee and co-workers reveal a cellular mechanism that protects male germ cells from heat stress. The researchers show that brief exposure of mouse testes to the core body temperature induces the assembly of stress granules (SGs; non-membranous cytoplasmic particles that contain translationally inert messenger ribonucleoproteins) in male germ cells. DAZL, a germ cell-specific translational regulator, translocates to SGs upon heat stress, they report, and is essential for their assembly. Importantly, DAZL-containing SGs sequester specific signalling molecules, such as RACK1 (receptor for activated protein kinase C), thereby blocking the apoptotic MAPK pathway. Together, these results suggest that DAZL is an essential component of SGs and that SGs prevent male germ cells from undergoing apoptosis upon heat stress.
Plus…
New for this year: poster articles!
Cilia in vertebrate development and disease
Oh and Katsanis provide a snapshot of the structure, function and distribution of the vertebrate cilium and of the pathologies that are associated with its dysfunction.
See the Development at a Glance poster article on p. 443
A taste of TGFβ in Tuscany
The recent FASEB Summer Research Conference entitled ‘The TGFβ Superfamily: Signaling in Development and Disease’ was held in August, 2011 in the spectacular setting of Il Ciocco, Lucca, amidst the olive trees in Tuscany, Italy. Here, Hata and Brivanlou review this meeting and highlight the recent advances that have been made in our understanding of the transforming growth factor-β (TGFβ) signaling pathway.
See the Meeting Review on p. 449
Regulation of DNA replication during development
During development, DNA replication is coordinated with cell proliferation and is regulated uniquely in specific cell types and organs. Here, Nordman and Orr-Weaver highlight recent advances and technologies that have provided us with new insights into the developmental regulation of DNA replication.
See the Review on p. 455
 (1 votes)
(1 votes)
 Loading...
Loading...




 (No Ratings Yet)
(No Ratings Yet) (1 votes)
(1 votes)




 Conference season is about to kick off, so here are a few registration dates for various meetings you might want to attend. If you know of any others, please leave a comment below. And of course keep checking the
Conference season is about to kick off, so here are a few registration dates for various meetings you might want to attend. If you know of any others, please leave a comment below. And of course keep checking the