Here are the highlights from the current issue of Development:
Skin-deep dermal niches
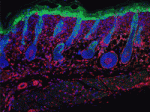
Hair follicle formation in the epidermis depends on signals from the underlying dermis and normally only occurs during late embryonic and early neonatal life. However, epidermal activation of β-catenin can induce follicle formation in adult mouse skin. One possible explanation for this observation is that epidermal cues can reprogram adult dermis to a neonatal state. On p. 5189, Fiona Watt and co-workers investigate this possibility by examining dermal fibroblasts from adult and neonatal mice. The researchers show that the gene expression profile of adult dermal fibroblasts isolated from skin in which β-catenin has induced ectopic follicles resembles that of neonatal fibroblasts rather than that of fibroblasts isolated from uninduced adult skin. This dermal reprogramming seems to originate within a specific subpopulation of fibroblasts near the hair follicle junctional zone and results in fibroblast proliferation and extracellular matrix remodelling. Together, these results suggest that the adult dermis is an unexpectedly plastic tissue, and that epidermal stem cells and their dermal niche exist in a state of dynamic interdependence.
Self-duplicating escorts for germ cells
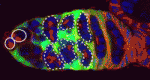
During Drosophila egg development, differentiated germ cells, which are produced by germline stem cells (GSCs), are accompanied by escort cells (ECs) to the centre of the germarium, where the germ cells form egg chambers. It has been proposed that these ECs are generated by a population of escort stem cells, but Ting Xie and co-workers now overturn this idea (p. 5087). The researchers show that ECs undergo slow turnover and that lost cells are replaced by self-duplication rather than by stem cell division. Using fluorescent markers, they show that ECs extend elaborate cellular processes that interact with differentiated germ cells and that these processes are missing when GSC differentiation is blocked. Conversely, disruption of Rho function in ECs, which disrupts the formation of EC processes, leads to the accumulation of ill-differentiated single germ cells and the gradual loss of ECs. These findings reveal a mutual dependence between ECs and differentiated GSC progeny, and suggest that self-maintained ECs form a niche that controls GSC lineage differentiation.
Lens invagination Shrooms along
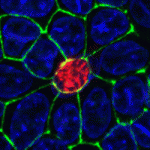
Epithelial invagination, a common feature of embryogenesis, involves coordinated modulation of individual cell cytoskeletons. For example, during eye development, lens pit invagination, which is accompanied by a columnar-to-conical cell shape change (termed apical constriction, AC), is dependent on the cytoskeletal protein Shroom3. Now, through experiments in chick and mouse embryos and in MDCK cells, Richard Lang and colleagues provide new insights into how Shroom3 drives AC and lens invagination (see p. 5177). The researchers show that the activity of Rock1/2 (serine/threonine kinases that activate non-muscle myosin and that are activated by the Rho family GTPase RhoA) is required for lens invagination and that RhoA activity is required for Shroom3-induced AC. RhoA, when activated and targeted apically, is sufficient to induce AC, they report, and is essential for the apical localisation of Shroom3. Finally, they show that the RhoA guanine nucleotide exchange factor Trio is required for Shroom3-dependent AC. Thus, a Trio-RhoA-Shroom3 pathway is required for AC during lens pit invagination.
Merlin casts a spell over glial cell proliferation
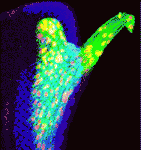
Glial cells perform many essential roles in the nervous system but how are their numbers controlled during development? Here (p. 5201), Venugopala Reddy and Kenneth Irvine report that glial cell proliferation in Drosophila is regulated by Hippo signalling, a conserved signalling pathway that controls organ growth and size. They show that Yorkie, the transcriptional co-activator of Hippo, is necessary for normal glial cell numbers and drives glial cell overproliferation when overexpressed. Yorkie activity in glial cells, they report, is controlled by a Merlin-Hippo signalling pathway; other upstream regulators of Hippo (for example, Fat) play no detectable role in glial cell proliferation. They also show that Yorkie affects glial cell proliferation by promoting the expression of the microRNA gene bantam, which, in turn, promotes Myc expression. Because homologues of Merlin, Yorkie and Myc are implicated in human glioma development, the authors suggest that the regulatory links identified here could represent a conserved pathway for the control of glial cell proliferation.
String-ing out stem cell homeostasis and aging
Stem cells contribute to tissue homeostasis throughout life by producing differentiating daughter cells. Consequently, a decline in stem cell proliferation is thought to be involved in tissue aging. But what regulates the cell cycle in stem cells? On p. 5079, Yukiko Yamashita and colleagues report that String (Stg), a homologue of the cell-cycle regulator Cdc25, controls stem cell maintenance, proliferation and aging in Drosophila testes. The researchers show that Stg is highly expressed in germline stem cells (GSCs) and in cyst stem cells (CySCs), the two stem cell types present in Drosophila testes, and that Stg is required for GSC and CySC maintenance and proliferation. Moreover, Stg expression declines with age in GSCs, and this decline is a major determinant of the age-related decline in GSC and CySC function. Notably, restoration of Stg activity reverses this age-associated phenotype but also leads to late-onset tumours. The researchers propose, therefore, that Stg/Cdc25 is a crucial regulator of stem cell function during tissue homeostasis and aging.
Brainy new role for Pax6
Successful development of the brain requires the tight regulation of sequential symmetric and asymmetric cell division. The molecular machinery that regulates the mode of cell division during mammalian brain development is poorly understood but now Magdalena Götz and colleagues show that the transcription factor Pax6, a known regulator of neurogenesis and proliferation, regulates both the orientation and mode of cell division in the mouse cerebral cortex (p. 5067). Using live imaging, the researchers show that, in the absence of Pax6, there is an increase in non-vertical cellular cleavage planes in the cerebral cortex. This phenotype, they report, seems to be mediated by the Pax6 target Spag5, which is a microtubule-associated protein. Moreover, long-term live imaging in vitro shows that Pax6-deficient progenitors generate daughter cells with asymmetric fates at higher frequencies than wild-type progenitors. From these and other data, the researchers propose that Pax6 plays a cell-autonomous role in the regulation of cortical progenitor cell division that is independent of apicobasal polarity and cell-cell interactions.
Plus…
Chromosome silencing mechanisms in X-chromosome inactivation: unknown unknowns
In this issue, fifty years after Mary Lyon proposed her X-chromosome inactivation (XCI) hypothesis, Neil Brockdorff discusses the molecular mechanisms of chromosome silencing during XCI, focusing on topics in which new findings are challenging the prevailing view. See the Review Article on p. 5057
Fifty years of X-inactivation research
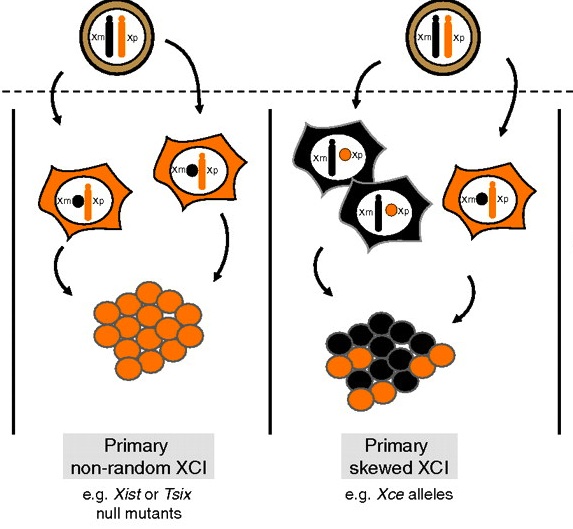
The third X-inactivation meeting ‘Fifty years of X-inactivation research’, which celebrated the fiftieth anniversary of Mary Lyon’s formulation of the X-inactivation hypothesis, was an EMBO workshop held in Oxford, UK, in July 2011. Gendrel and Heard review the results presented at the meeting and highlight some of the exciting progress that has been made in this field. See the Meeting Review on p. 5049
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (1 votes)
(1 votes) (No Ratings Yet)
(No Ratings Yet)










 Dates for your calendar
Dates for your calendar