Here are the research highlights from the current issue of Development:
Shaping up the Hippo pathway
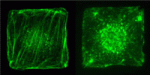
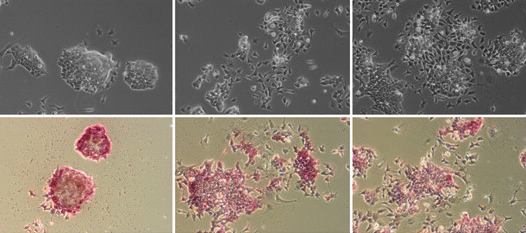
The Hippo pathway, which regulates cell proliferation, is regulated by cell density: low cell density induces weak Hippo signalling, leading to nuclear accumulation of the transcriptional co-activator Yap and the promotion of proliferation, whereas high cell density prevents nuclear accumulation of Yap and suppresses proliferation. The mechanisms by which cells detect density, however, are unknown. Here, on p. 3907, Hiroshi Sasaki and colleagues show that cell morphology plays a key role in regulating the Hippo pathway. The researchers show that manipulation of NIH3T3 cell morphology, by culture on fabricated microdomains, regulates the subcellular localisation of Yap. These changes in cell morphology, they report, lead to changes in actin stress fiber quantities and the subsequent regulation of Yap phosphorylation and localisation. Finally, the researchers show that stress fibers regulate Yap upstream of, or at the level of, the protein kinase Lats. The researchers thus propose that a cell morphology-based mechanism, mediated by stress fibers, cooperates with a cell adhesion-based mechanism to achieve density-dependent control of cell proliferation.
Vreteno: a novel protein in germline piRNA biogenesis
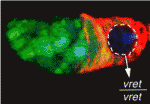
In Drosophila, Piwi-interacting RNAs (piRNAs) preserve genome integrity in the germline by silencing mobile genetic elements, such as transposons. On p. 4039, Ruth Lehmann and co-workers report the identification of Vreteno, a novel gonad-specific protein that is essential for germline development and primary piRNA biogenesis in Drosophila. The researchers demonstrate that vreteno (vret), which was identified in a screen for maternal-effect mutations affecting oocyte polarity, is essential for germline development. They further show that Vret, which contains two Tudor domains, interacts with Piwi and Aubergine to regulate their stability and subcellular localisation. Using microarray analyses, they confirm that vret regulates transposon silencing in both the germline and somatic tissues of the Drosophila gonad. Finally, the authors report, in the absence of Vret, Piwi-bound piRNAs are lost, whereas piRNAs can engage in Aubergine- and Argonaute 3-dependent `ping-pong’ amplification. The authors thus suggest that Vreteno regulates transposon silencing by acting at the early stages of primary piRNA processing.
Joining forces: PCP and apical-basal polarity
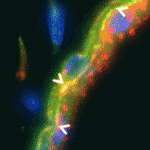
Cell polarity can be defined in terms of the polarity of a cell with respect to others in the same plane (planar cell polarity; PCP), or in terms of polarity based on the subcellular localisation of cell structures, proteins or domains (apical-basal polarity; ABP). The extent to which these polarity pathways are linked, however, is unclear. Here, Janet Heasman and colleagues investigate interactions between the PCP protein Vangl2 and the ABP component aPKC in Xenopus oocytes (p. 3989). The researchers show that Vangl2 is enriched animally in subcortical islands, where it interacts with vesicle associated membrane protein 1 (VAMP1) and acetylated microtubules. The distribution of these islands and the microtubule cytoskeleton, they report, is dependent on aPKC. Importantly, the researchers show that both maternal Vangl2 and aPKC are required to establish asymmetries in the oocyte and early embryo. These data highlight important links between the PCP and ABP pathways, suggesting that Vangl2 and aPKC are part of a common network that influences oocyte and embryo patterning.
Snail enhancers step out of the shade

The expression of critical developmental genes can be regulated by multiple cis-regulatory modules (CRMs), and it has been suggested that remote CRMs are redundant to promoter proximal CRMs. But what is the function of these multiple CRMs and are they truly redundant? To answer this question, Angelike Stathopoulos and co-workers (p. 4075) examine two CRMs from the Drosophila snail gene locus and show that these CRMs interact in a non-additive manner to regulate snail expression. The researchers demonstrate that the CRMs drive distinct patterns of gene expression in early embryos. Furthermore, they report, the distal CRM acts to limit the expanded expression domain of the proximal CRM, whereas the proximal CRM serves to `dampen’ the levels of expression driven by the distal CRM. Importantly, the CRMs are not functionally equivalent; only the distal CRM is required in snail transgenes to rescue snail mutants. Thus, the authors propose, complex interactions between CRMs are required for fine-tuning the patterns and levels of snail expression during development.
Ciliogenesis: arrested development at the node

The rotation of cilia on cells within the node of mammalian embryos generates a leftward fluid flow that establishes left-right asymmetry. But what regulates ciliogenesis at the node? Here (p. 3915), Yuji Mishina and colleagues show that cell cycle arrest, mediated by bone morphogenetic protein (BMP) signalling, is required in node cells to trigger nodal ciliogenesis in mice. The authors show that epiblast-specific deletion of Acvr1, which encodes a BMP type 1 receptor, results in abnormal left-right patterning in early embryos; the node forms in these mutants but nodal ciliogenesis is compromised. Using Acvr1-deficient mouse embryonic fibroblasts, they further demonstrate that BMP signalling through ACVR1 positively regulates p27Kip1 stability and phosphorylation, which in turn maintains quiescence and allows the formation of primary cilia. Importantly, the researchers report, p27Kip1 is present and phosphorylated in quiescent nodal cells, whereas the corresponding cells in Acvr1 mutants are proliferative and show reduced p27Kip1 expression and phosphorylation. These studies provide valuable insight into the mechanisms by which primary cilia form at the node.
Distinct roles for Nodal and Cripto in stem cells
Extra-embryonic endoderm stem (XEN) cells can be derived from the mouse primitive endoderm, which gives rise to two extra-embryonic tissues: the visceral endoderm (VE) and the parietal endoderm. However, despite displaying many characteristics of primitive endoderm, XEN cells only contribute effectively to parietal endoderm in mouse chimeras. Here, Michael Shen and co-workers study the differentiation of XEN cells in response to Nodal, a member of the TGFβ superfamily, and Cripto, a Nodal co-receptor (p. 3885). Importantly, the researchers show that XEN cells treated with either Nodal or Cripto display an up-regulation of VE markers and contribute to VE in chimeric embryos. Notably, they report, the response of XEN cells to Nodal and Cripto differs: the response to Nodal is blocked by treatment with an Alk4/Alk5/Alk7 kinase inhibitor, whereas the response to Cripto is unaffected, suggesting that Cripto can act independently of these receptors’ activity. These findings provide key insights into visceral endoderm specification and define distinct pathways for Nodal and Cripto during cell differentiation.
Plus…
Mechanisms of thymus organogenesis and morphogenesis
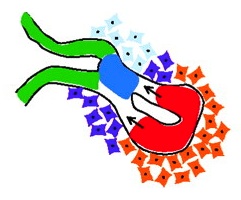
The thymic microenvironment supports T cell development and regeneration and, as reviewed by Gordon and Manley, recent studies have made significant progress in identifying the mechanisms that control the specification, early organogenesis and morphogenesis of the thymus. See the Review article on p. 3865
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...
 Survey
Survey

 (No Ratings Yet)
(No Ratings Yet) (1 votes)
(1 votes) While it may look like an African violet, this is actually a staining of the four-cell stage of a slipper limpet (Crepidula fornicata) just about to cleave to the eight-cell stage. This image, taken by Anna Franz of the University of Oxford, was one of the candidates in the
While it may look like an African violet, this is actually a staining of the four-cell stage of a slipper limpet (Crepidula fornicata) just about to cleave to the eight-cell stage. This image, taken by Anna Franz of the University of Oxford, was one of the candidates in the