July in preprints
Posted by the Node, on 9 August 2022
Welcome to our monthly trawl for developmental biology (and related) preprints.
The preprints this month are hosted on bioRxiv, arXiv and preprints.org – use these links to get to the section you want.
- Patterning & signalling
- Morphogenesis & mechanics
- Genes & genomes
- Stem cells, regeneration & disease modelling
- Plant development
- Evo-devo
Developmental biology
| Patterning & signalling
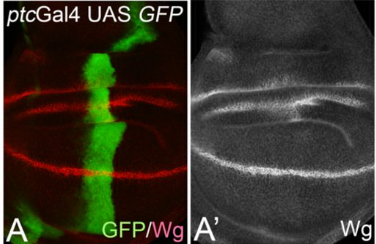
Cis-inhibition suppresses basal Notch signalling during sensory organ precursor selection
Tobias Troost, Udi Binshtok, David Sprinzak, Thomas Klein
Innexin function dictates the spatial relationship between distal somatic cells in the Caenorhabditis elegans gonad without impacting the germline stem cell pool
Theadora Tolkin, Ariz Mohammad, Todd Starich, Ken C. Q. Nguyen, David H. Hall, Tim Schedl, E. Jane Albert Hubbard, David Greenstein
Fibronectin deficiency in newborn mice leads to cyst formation in the kidney
Kristina Hermann, Silke Seibold, Kathrin Skoczynski, Bjoern Buchholz, Ernst R. Tamm, Leonie Herrnberger-Eimer
Canonical Wnt Signaling Maintains Human Mesenchymal Progenitor Cell Multipotency During Adipose Tissue Development
Zinger Yang Loureiro, Shannon Joyce, Javier Solivan-Rivera, Anand Desai, Pantos Skritakis, Qin Yang, Tiffany DeSouza, Tammy Nguyen, Ormond A MacDougald, Silvia Corvera
Evidence for intercellular bridges and radial patterning of meiotic initiation in the human fetal ovary
Bikem Soygur, Amber Derpinghaus, Gerald R. Cunha, Laurence S. Baskin, Diana J Laird
The C. elegans gonadal sheath Sh1 cells extend asymmetrically over a differentiating germ cell population in the proliferative zone
Xin Li, Noor Singh, Camille Miller, India Washington, Bintou Sosseh, Kacy Lynn Gordon
Foxi3 Suppresses Signaling Center Fate and is Necessary for the Early Development of Mouse Teeth
Isabel Mogollón, Niko Kangasniemi, Jacqueline Emmanuel Moustakas-Verho, Laura Ahtiainen
A Notch-dependent transcriptional mechanism controls expression of temporal patterning factors in Drosophila medulla
Alokananda Ray, Xin Li
Stretch Regulates Alveologenesis and Homeostasis Via Mesenchymal Gαq/11-Mediated TGFβ2 Activation
Amanda T Goodwin, Alison E John, Chitra Joseph, Anthony Habgood, Amanda L Tatler, Katalin Susztak, Matthew Palmer, Stefan Offermanns, Neil C Henderson, R Gisli Jenkins
A PAK kinase family member and the Hippo/Yorkie pathway modulate WNT signaling to functionally integrate body axes during regeneration
Viraj Doddihal, Frederick G. Mann Jr., Eric Ross, Sean A. McKinney, Alejandro Sánchez Alvarado
Germline protein, Cup, non-cell autonomously limits migratory cell fate in Drosophila oogenesis
Banhisikha Saha, Sayan Acharjee, Gaurab Ghosh, Purbasa Dasgupta, Mohit Prasad
CXCR7 promotes foetal myoblast fusion at muscle fiber tips independently of Myomaker via a ß1integrin-EGFR-dependent mechanism
Sonya Nassari, Cédrine Blavet, Delphine Duprez, Claire Fournier-Thibault
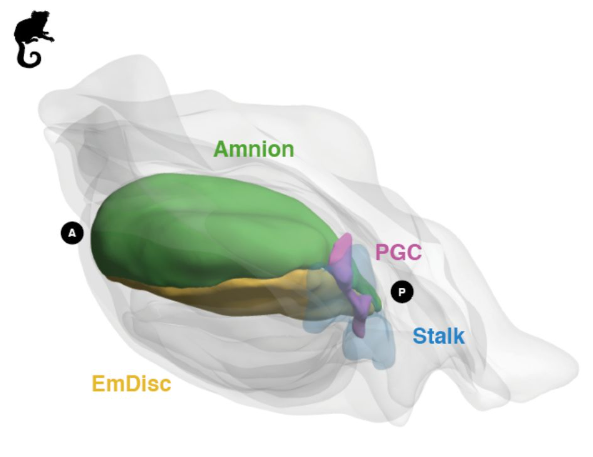
Origin and segregation of the human germline
Aracely Castillo-Venzor, Christopher A. Penfold, Michael D. Morgan, Walfred W. C. Tang, Toshihiro Kobayashi, Frederick C. K. Wong, Sophie Bergmann, Erin Slatery, Thorsten E. Boroviak, John C. Marioni, M. Azim Surani
Ordered deployment of distinct ciliary beating machines in growing axonemes of vertebrate multiciliated cells
Chanjae Lee, Yun Ma, Fan Tu, John B. Wallingford
AKT1-FOXO4 AXIS RECIPROACLLY REGULATES HEMOCHORIAL PLACENTATION
Keisuke Kozai, Ayelen Moreno-Irusta, Khursheed Iqbal, Mae-Lan Winchester, Regan L. Scott, Mikaela E. Simon, Masanaga Muto, Marc R. Parrish, Michael J. Soares
A mutant bacterial O-GlcNAcase visualizes a progressive decline of protein O-GlcNAcylation in early Drosophila embryos critical for neurodevelopment
Yaowen Zhang, Dandan Wang, Haibin Yu, Xiaoyun Lei, Yang Meng, Na Zhang, Fang Chen, Lu Lv, Qian Pan, Hongtao Qin, Zhuohua Zhang, Daan M.F. van Aalten, Kai Yua
The extracellular matrix protein fibronectin modulates metanephric kidney development
Kathrin Skoczynski, Andre Kraus, Maike Büttner-Herold, Kerstin Amann, Mario Schiffer, Kristina Hermann, Leonie Herrnberger, Ernst R. Tamm, Bjoern Buchholz
A conserved role of Hippo signaling in initiation of the first lineage specification event across mammals
Claudia Gerri, Afshan McCarthy, Gwen Mei Scott, Marius Regin, Sophie Brumm, Claire S. Simon, Janet Lee, Cristina Montesinos, Caroline Hassitt, Sarah Hockenhull, Daniel Hampshire, Kay Elder, Phil Snell, Leila Christie, Ali A. Fouladi-Nashta, Hilde Van de Velde, Kathy K. Niakan
Loss of growth differentiation factor 9 causes an arrest of early folliculogenesis in zebrafish – a novel insight into its action mechanism
Weiting Chen, Yue Zhai, Bo Zhu, Kun Wu, Yuqin Fan, Xianqing Zhou, Lin Liu, Wei Ge
Nonlinear effect of light intensity on normal axial development of rhesus monkeys
Ying-Zhou Hu, Hua Yang, Jing Wu, Hao Li, Long-Bao Lv, Zhu Zhu, Lu-Yao Zhou, Yu-Hua Zhang, Fang-Fang Yan, Shu-Han Fan, Cheng-Yu Li, Shu-Xiao Wang, Jian-Ping Zhao, Qiang Qi, Chang-Bing Huang, Xin-Tian Hu
Specification of the endocrine primordia controlling insect moulting and metamorphosis by the JAK/STAT signalling pathway
Mar García-Ferrés, Carlos Sánchez-Higueras, Jose Manuel Espinosa-Vázquez, James C-G Hombría
DMRT1 regulation of TOX3 modulates expansion of the gonadal steroidogenic cell lineage
Martin A. Estermann, Andrew T. Major, Craig A. Smith
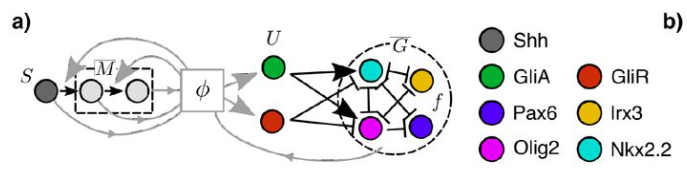
Optimal control of gene regulatory networks for morphogen-driven tissue patterning
A. Pezzotta, J. Briscoe
Early pre-neural serotonin modulates balance of late monoamines and behavioral patterns in fish model system
Evgeny Ivashkin, Stefan Spulber, Andrei Zinovyev, Takashi Yoshitake, Shimako Yoshitake, Olga Kharchenko, Marina Yu. Khabarova, Spyridon Theofilopoulos, Jan Kehr, Ernest Arenas, Sandra Ceccatelli, Elena E. Voronezhskaya, Igor Adameyko
mTORC1 is required for differentiation of germline stem cells in the Drosophila melanogaster testis
Marie Clémot, Cecilia D’Alterio, Alexa Kwang, D. Leanne Jones
Dissecting the roles of Expansion/Rebuf and the chitin synthase Krotzkopf Verkehrt in chitin deposition in Drosophila
Ettore De Giorgio, Panagiotis Giannios, M. Lluisa Espinàs, Marta Llimargas
| Morphogenesis & mechanics
Reciprocal regulation between cell mechanics and ZO-1 guides tight junction assembly and epithelial morphogenesis
Alexis J. Haas, Ceniz Zihni, Susanne M. Krug, Riccardo Maraspini, Tetsuhisa Otani, Mikio Furuse, Alf Honigmann, Maria Balda, Karl Matter
Afadin and zyxin contribute to coupling between cell junctions and contractile actomyosin networks during apical constriction
Mark M. Slabodnick, Sophia C. Tintori, Mangal Prakash, Christopher D. Higgins, Alicia H. Chen, Timothy D. Cupp, Terrence Wong, Emily Bowie, Florian Jug, Bob Goldstein
Growth anisotropy of the extracellular matrix drives mechanics in a developing organ
Stefan Harmansa, Alexander Erlich, Christophe Eloy, Giuseppe Zurlo, Thomas Lecuit
Exocyst Inactivation in Urothelial Cells Disrupts Autophagy and Activates non-canonical NF-κB
Michael A. Ortega, Ross K. Villiger, Malia Harrison-Chau, Suzanna Lieu, Kadee-Kalia Tamashiro, Amanda J. Lee, Brent A. Fujimoto, Geetika Y. Patwardhan, Joshua Kepler, Ben FogelgrenMichael A. Ortega, Ross K. Villiger, Malia Harrison-Chau, Suzanna Lieu, Kadee-Kalia Tamashiro, Amanda J. Lee, Brent A. Fujimoto, Geetika Y. Patwardhan, Joshua Kepler, Ben Fogelgren
Imaginal disc growth factors are Drosophila Chitinase-like Proteins with roles in morphogenesis and CO2 response
Anne Sustar, Liesl Strand, Sandra Zimmerman, Celeste Berg
Frem1 activity regulated by Sonic Hedgehog signaling in the cranial neural crest mesenchyme guides midfacial morphogenesis
Matthew T. McLaughlin, Miranda R. Sun, Tyler G. Beames, Austin C. Steward, Joshua W. M. Theisen, Hannah M. Chung, Joshua L. Everson, Ivan P. Moskowitz, Michael D. Sheets, Robert J. Lipinski
Actomyosin contractility in olfactory placode neurons opens the skin epithelium to form the nostril
Marion Baraban, Clara Gordillo Pi, Isabelle Bonnet, Jean-François Gilles, Camille Lejeune, Mélody Cabrera, Florian Tep, Marie Anne Breau
Genetic and geometric heredity interact to drive polarized flow in the Drosophila embryo
Emily Gehrels, Bandan Chakrabortty, Matthias Merkel, Thomas Lecuit
Micropatterned Organoids Enable Modeling of the Earliest Stages of Human Cardiac Vascularization
Oscar J. Abilez, Huaxiao Yang, Lei Tian, Kitchener D. Wilson, Evan H. Lyall, Mengcheng Shen, Rahulkumar Bhoi, Yan Zhuge, Fangjun Jia, Hung Ta Wo, Gao Zhou, Yuan Guan, Bryan Aldana, Detlef Obal, Gary Peltz, Christopher K. Zarins, Joseph C. Wu
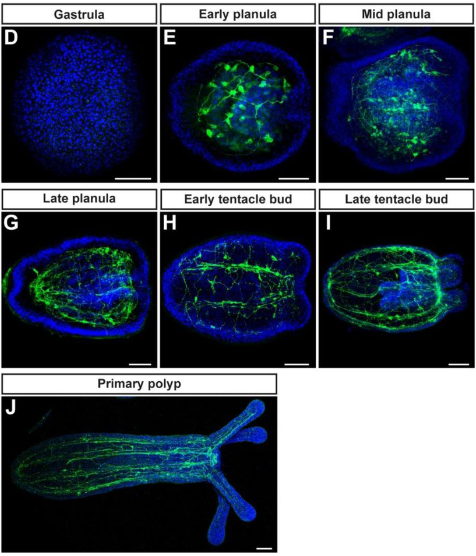
NvPrdm14d-expressing neural progenitor cells contribute to non-ectodermal neurogenesis in Nematostella vectensis
Quentin I. B. Lemaître, Natascha Bartsch, Ian U. Kouzel, Henriette Busengdal, Gemma Sian Richards, Patrick R. H. Steinmetz, Fabian Rentzsch
Human-specific progenitor sub-domain contributes to extended neurogenesis and increased motor neuron production
Sumin Jang, Elias Gunmit, Hynek Wichterle
CITED2 Is A Conserved Regulator Of Deep Hemochorial Placentation
Marija Kuna, Pramod Dhakal, Khursheed Iqbal, Esteban M. Dominguez, Lindsey N. Kent, Masanaga Muto, Ayelen Moreno-Irusta, Keisuke Kozai, Kaela M. Varberg, Hiroaki Okae, Takahiro Arima, Henry M. Sucov, Michael J. Soares
Spatial consistency of cell growth direction during organ morphogenesis requires CELLULOSE-SYNTHASE INTERACTIVE1
Corentin Mollier, Joanna Skrzydeł, Dorota Borowska-Wykret, Mateusz Majda, Mateusz Dulski, Antoine Fruleux, Roman Wrzalik, Richard S. Smith, Françoise Monéger, Dorota Kwiatkowska, Arezki Boudaoud
Single Cell Epigenetics Reveal Cell-Cell Communication Networks in Normal and Abnormal Cardiac Morphogenesis
Sanjeev S. Ranade, Sean Whalen, Ivana Zlatanova, Tomohiro Nishino, Benjamin van Soldt, Lin Ye, Angelo Pelonero, Langley Grace Wallace, Yu Huang, Michael Alexanian, Arun Padmanabhan, Barbara Gonzalez-Teran, Pawel Przytycki, Mauro W. Costa, Casey A. Gifford, Brian L. Black, Katherine S. Pollard, Deepak Srivastava
Characterization of the human fetal rete region by single cell transcriptional analysis of gonads and mesonephros/epididymis
Jasin Taelman, Sylwia M. Czukiewska, Ioannis Moustakas, Yolanda W. Chang, Sanne Hillenius, Talia van der Helm, Hailiang Mei, Xueying Fan, Susana M. Chuva de Sousa Lopes
A molecular mechanism for membrane chaperoning by a late embryogenesis abundant protein
Xiao-Han Li, Conny W.H. Yu, Natalia Gomez-Navarro, Viktoriya Stancheva, Hongni Zhu, Cristina Guibao, Andal Murthy, Boer Xie, Michael Wozny, Benjamin Leslie, Marcin Kaminski, Ketan Malhotra, Christopher M. Johnson, Martin Blackledge, Balaji Santhanam, Douglas R. Green, Junmin Peng, Wei Liu, Jinqing Huang, Elizabeth A. Miller, Stefan M.V. Freund, M. Madan Babu
| Genes & genomes
Obox4 secures zygotic genome activation upon loss of Dux
Youjia Guo, Tomohiro Kitano, Kensaku Murano, Ten D. Li, Akihiko Sakashita, Hirotsugu Ishizu, Masayuki Sato, Haruhiko Siomi
Transcription factors regulating the fate and developmental potential of a multipotent progenitor in C. elegans
Evan M. Soukup, Jill C. Bettinger, Laura D. Mathies
Context-dependent transcriptional remodeling of TADs during differentiation
Sanjay Chahar, Yousra Ben Zouari, Hossein Salari, Anne M Molitor, Dominique Kobi, Manon Maroquenne, Cathie Erb, Audrey Mossler, Nezih Karasu, Daniel Jost, Tom Sexton
BmHen1 plays an essential role in the regulation of eupyrene sperm development in Bombyx mori
Xu Yang, Dongbin Chen, Shirui Zheng, Meiyan Yi, Zulian Liu, Yongjian Liu, Dehong Yang, Yujia Liu, Linmeng Tang, Chenxu Zhu, Yongping Huang
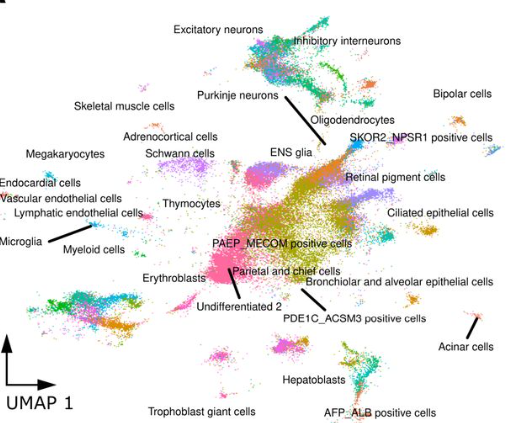
Embryoid bodies facilitate comparative analysis of gene expression in humans and chimpanzees across dozens of cell types
Kenneth A Barr, Katherine L Rhodes, Yoav Gilad
The logic of native enhancer-promoter compatibility and cell-type-specific gene expression variation
Takeo Narita, Yoshiki Higashijima, Sinan Kilic, Elina Maskey, Katrin Neumann, Chunaram Choudhary
Histone demethylome map reveals combinatorial gene regulatory functions in embryonic stem cells
Yogesh Kumar, Pratibha Tripathi, Pushkar Dakle, Majid Mehravar, Varun K. Pandey, Michael J. Bullen, Zhongming Zhang, Dhaval Hathiwala, Marc Kerenyi, Andrew Woo, Alireza Ghamari, Alan B. Cantor, Lee H. Wong, Jonghwan Kim, Kimberly Glass, Guo-Cheng Yuan, Luca Pinello, Stuart H. Orkin, Partha Pratim Das
Transcription of Murine Endogenous Retrovirus MERVL Is Required for Progression of Development in Early Preimplantation Embryos
Akihiko Sakashita, Tomohiro Kitano, Hirotsugu Ishizu, Youjia Guo, Harumi Masuda, Masaru Ariura, Kensaku Murano, Haruhiko Siomi
Lineage-specific, fast-evolving GATA-like gene regulates zygotic gene activation to promote endoderm specification and pattern formation in the Theridiidae spider
Sawa Iwasaki-Yokozawa, Ryota Nanjo, Yasuko Akiyama-Oda, Hiroki Oda
Plap-1/Aspn lineage tracing and single-cell transcriptomics reveals cellular dynamics in the periodontal ligament
Tomoaki Iwayama, Mizuho Iwashita, Kazuya Miyashita, Hiromi Sakashita, Shuji Matsumoto, Kiwako Tomita, Phan Bhongsatiern, Tomomi Kitayama, Kentaro Ikegami, Takashi Shimbo, Katsuto Tamai, Masanori A Murayama, Shuhei Ogawa, Yoichiro Iwakura, Satoru Yamada, Lorin E Olson, Masahide Takedachi, Shinya Murakami
Stepwise progression of β-selection during T cell development as revealed by histone deacetylation inhibition
Anchi S Chann, Mirren Charnley, Lucas M. Newton, Andrea Newbold, Florian Wiede, Tony Tiganis, Patrick O Humbert, Ricky W Johnstone, Sarah M Russell
Efficient Human Germ Cell Specification from Stem Cells via Combinatorial Expression of Transcription Factors
Christian Kramme, Merrick Pierson Smela, Bennett Wolf, Patrick R. Fortuna, Garyk Brixi, Kalyan Palepu, Edward Dong, Jessica Adams, Suhaas Bhat, Sabrina Koseki, Emma Tysinger, Teodora Stan, Richie E. Kohman, Songlei Liu, Mutsumi Kobayashi, Toshi Shioda, George M. Church, Pranam Chatterjee
Activating and repressing gene expression between chromosomes during stochastic fate specification
Elizabeth A. Urban, Chaim Chernoff, Kayla Viets Layng, Jeong Han, Caitlin Anderson, Daniel Konzman, Robert J. Johnston Jr.
Young transposable elements rewired gene regulatory networks in human and chimpanzee hippocampal intermediate progenitors
Sruti Patoori, Samantha M. Barnada, Christopher Large, John I. Murray, Marco Trizzino
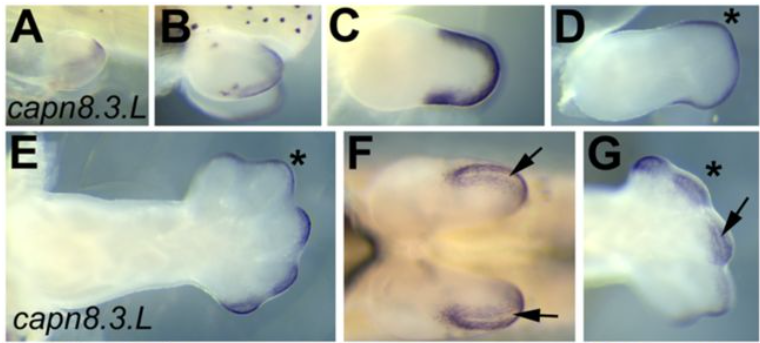
Gene expression analysis of the Xenopus laevis early limb bud proximodistal axis
D.T. Hudson, J. S. Bromell, R.C. Day, T McInnes, J.M. Ward, C.W. Beck
A New CUT&RUN Low Volume-Urea (LoV-U) protocol uncovers Wnt/β-catenin tissue-specific genomic targets
Gianluca Zambanini, Anna Nordin, Mattias Jonasson, Pierfrancesco Pagella, Claudio Cantù
Sociosexual behavior requires both activating and repressive roles of Tfap2e/AP- 2ε in vomeronasal sensory neurons
Jennifer M. Lin, Tyler A. Mitchell, Megan Rothstein, Alison Pehl, Ed Zandro M. Taroc, Raghu Ram Katreddi, Katherine E. Parra, Damian G. Zuloaga, Marcos Simoes-Costa, Paolo E. Forni
Control of neuronal terminal differentiation through cell context-dependent CFI-1/ARID3 functions
Yinan Li, Jayson J. Smith, Filipe Marques, Anthony Osuma, Hsin-Chiao Huang, Paschalis Kratsios
Changes of chromosomal architecture before establishment of chromosome territories revealed by recurrence plot reconstruction
Yuki Kitanishi, Hiroki Sugishita, Yukiko Gotoh, Yoshito Hirata
Single cell analysis of lymphatic endothelial cell fate specification and differentiation during zebrafish development
Lin Grimm, Elizabeth Mason, Oliver Yu, Stefanie Dudczig, Virginia Panara, Tyrone Chen, Neil I. Bower, Scott Paterson, Kazuhide Okuda, Maria Rondon Galeano, Sakurako Kobayashi, Anne Senabouth, Anne K. Lagendijk, Joseph Powell, Kelly A. Smith, Katarzyna Koltowska, Benjamin M. Hogan
Novel cell- and stage-specific transcriptional signatures defining Drosophila neurons, glia and hemocytes
Rosy Sakr, Pierre B. Cattenoz, Alexia Pavlidaki, Laura Ciapponi, Marta Marzullo, Nivedita Hariharan, Tina Mukherjee, Angela Giangrande
High Sox2 expression predicts taste lineage competency of lingual progenitors in vitro
Lauren A. Shechtman, Jennifer K. Scott, Eric D. Larson, Trevor J. Isner, Bryan J. Johnson, Dany Gaillard, Peter J. Dempsey, Linda A. Barlow
Nuclear architecture protein Distal antenna balances genome-binding and phase-separation properties to regulate neuroblast competence
Gillie Benchorin, Maggie Jiaqi Li, Richard Jangwon Cho, Yuxin Hu, Minoree Kohwi
Germ-cell specific eIF4E1B regulates maternal RNA translation to ensure zygotic genome activation
Guanghui Yang, Qiliang Xin, Iris Feng, Jurrien Dean
RNA-binding protein Elavl1/HuR is required for maintenance of cranial neural crest specification
Erica J. Hutchins, Shashank Gandhi, Jose Chacon, Michael L. Piacentino, Marianne E. Bronner
Diverse logics and grammar encode notochord enhancers
Benjamin P Song, Michelle F Ragsac, Krissie Tellez, Granton A Jindal, Jessica L Grudzien, Sophia H Le, Emma K Farley
linc-mipep and linc-wrb encode micropeptides that regulate chromatin accessibility in vertebrate-specific neural cells
Valerie A. Tornini, Ho-Joon Lee, Liyun Miao, Yin Tang, Sarah E. Dube, Timothy Gerson, Valeria J. Schmidt, Katherine Du, Manik Kuchroo, François Kroll, Charles E. Vejnar, Ariel A. Bazzini, Smita Krishnaswamy, Jason Rihel, Antonio J. Giraldez
Histone 4 lysine 5/12 acetylation provides a plasticity code with epigenetic memory of environmental exposure
Michael S. Werner, Tobias Loschko, Thomas King, Tobias Theska, Mirita Franz-Wachtel, Boris Macek, Ralf J. Sommer
Molecular Underpinnings and Environmental Drivers of Spontaneous Loss of Heterozygosity in Drosophila Intestinal Stem Cells
Lara Al zouabi, Marine Stefanutti, Nick Riddiford, Natalia Rubanova, Mylène Bohec, Nicolas Servant, Allison Bardin
Symbiosis-driven development in an early branching metazoan
Aki H. Ohdera, Justin Darymple, Viridiana Avila-Magaña, Victoria Sharp, Kelly Watson, Mark McCauley, Bailey Steinworth, Erika M. Diaz-Almeyda, Sheila A. Kitchen, Angela Z. Poole, Anthony Bellantuono, Sajeet Haridas, Igor V. Grigoriev, Lea Goentoro, Elizabeth Vallen, David M. Baker, Todd C. LaJeunesse, Sandra Loesgen, Mark Q. Martindale, Matthew DeGennaro, William K. Fitt, Mónica Medina
| Stem cells, regeneration & disease modelling
Brain natriuretic peptide improves heart regeneration after infarction by stimulating cardiomyocyte renewal
Anne-Charlotte Bon-Mathier, Tamara Déglise, Stéphanie Rignault-Clerc, Christelle Bielmann, Lucia Mazzolai, Nathalie Rosenblatt-Velin
Identification of a multipotent lung progenitor for lung regeneration
Chava Rosen, Elias Shetzen, Irit Milman -Krentsis, Ran Orgad, Xiaohua Su, Raj Yadav, Michal Shemesh, Adi Biram, Ziv Shulman, Smadar Eventov-Friedman, Mukesh Maharjan, Yuan Qi, Jing Wang, Yair Reisner
Chemical induction of gut β-like-cells by combined FoxO1/Notch inhibition as a glucose-lowering treatment for diabetes
Takumi Kitamoto, Yun-Kyoung Lee, Nishat Sultana, Wendy M. McKimpson, Hitoshi Watanabe, Wen Du, Jason Fan, Bryan Diaz, Hua V. Lin, Rudolph L. Leibel, Sandro Belvedere, Domenico Accili
Localized heterochrony integrates overgrowth potential of oncogenic clones
Nicola Blum, Matthew P. Harris
Multi-chamber cardioids unravel human heart development and cardiac defects
Clara Schmidt, Alison Deyett, Tobias Ilmer, Aranxa Torres Caballero, Simon Haendeler, Lokesh Pimpale, Michael A. Netzer, Lavinia Ceci Ginistrelli, Martina Cirigliano, Estela Juncosa Mancheno, Daniel Reumann, Katherina Tavernini, Steffen Hering, Pablo Hofbauer, Sasha Mendjan
Aberrant extracellular matrix and cardiac development in models lacking the PR-DUB component ASXL3
BT McGrath, YC Tsan, S Salvi, N Ghali, DM Martin, M Hannibal, CE Keegan, A Helms, A Srivastava, SL Bielas
Imp is required for timely exit from quiescence in Drosophila type II neuroblasts
Jordan A. Munroe, Mubarak H. Syed, Chris Q. Doe
leptin b and its regeneration enhancer illustrate the regenerative features of zebrafish hearts
Kwangdeok Shin, Ian J. Begeman, Jingli Cao, Junsu Kang
Visualization of Retroplacental Clear Space Disruption in a Mouse Model of Placental Accreta
Andrew A. Badachhape, Prajwal Bhandari, Laxman Devkota, Mayank Srivastava, Eric A. Tanifum, Verghese George, Karin A. Fox, Chandrasekhar Yallampalli, Ananth V. Annapragada, Ketan B. Ghaghada
Multi-omics analyses identify transcription factor interplay in corneal epithelial fate determination and disease
Jos GA Smits, Dulce Lima Cunha, Jieqiong Qu, Nicholas Owen, Lorenz Latta, Nora Szentmary, Berthold Seitz, Lauriane N Roux, Mariya Moosajee, Daniel Aberdam, Simon J. van Heeringen, Huiqing Zhou
Neonatal hyperoxia induces sex-dependent pulmonary cellular and transcriptomic changes in an experimental mouse model of bronchopulmonary dysplasia
Sheng Xia, Lisandra Vila Ellis, Konner Winkley, Heather Menden, Sherry M. Mabry, Daniel Louiselle, Margaret Gibson, Elin Grundberg, Jichao Chen, Venkatesh Sampath
Variation in whole-body regeneration between Botrylloides morphs and species
Berivan Temiz, Megan J. Wilson
Autophagy slows the aging of Germline stem cells in Drosophila through modulation of E-cadherin
Nidhi Murmu, Bhupendra V. Shravage
Sex-bias in utero alters ovarian reserve but not uterine capacity in female offspring
Annika V Geijer-Simpson, Haidee Tinning, Tiago H C de Bem, Ioannis Tsagakis, Alysha S Taylor, Laura Hume, Lisa M Collins, Niamh Forde
Graft of cardiac progenitors in a pig model of right ventricular failure triggers myocardial epimorphosis, regeneration and protection of function
V Lambert, A Deleris, F Tibourtine, V Fouilloux, A Martin, P Bridge, E Aries, D Benoist, M Pucéat
Mapping fetal myeloid differentiation in airway samples from premature neonates with single-cell profiling
Holly Welfley, Ranjit Kylat, Nahla Zaghloul, Marilyn Halonen, Fernando D. Martinez, Mohamed Ahmed, Darren A. Cusanovich
Ethanol Exposure Perturbs Sea Urchin Development and Disrupts Developmental Timing
Nahomie Rodríguez-Sastre, Nicholas Shapiro, Dakota Y. Hawkins, Alexandra T. Lion, Monique Peyreau, Andrea E. Correa, Kristin Dionne, Cynthia A. Bradham
CLASP1 is essential for neonatal lung function and survival in mice
Ana L. Pereira, Tiago F. da Silva, Luísa T. Ferreira, Martine Jaegle, Marjon Buscop-van Kempen, Robbert Rottier, Wilfred F. J. van Ijcken, Pedro Brites, Niels Galjart, Helder Maiato
Specific Deletion of Axin1 Leads to Activation of β-Catenin/BMP Signaling Resulting in Fibular Hemimelia Phenotype in Mice
Rong Xie, Dan Yi, Qiang Jie, Qinglin Kang, Zeng Zhang, Zhenlin Zhang, Guozhi Xiao, Lin Chen, Liping Tong, Di Chen
Directed Differentiation of Human iPSCs to Functional Ovarian Granulosa-Like Cells via Transcription Factor Overexpression
Merrick Pierson Smela, Christian Kramme, Patrick Fortuna, Jessica Adams, Edward Dong, Mutsumi Kobayashi, Garyk Brixi, Emma Tysinger, Richie. E. Kohman, Toshi Shioda, Pranam Chatterjee, George M. Church
Odd skipped-related 1 controls the pro-regenerative response of Fibro-Adipogenic Progenitors
Georgios Kotsaris, Taimoor H. Qazi, Christian H. Bucher, Sophie Pöhle-Kronawitter, Vladimir Ugorets, William Jarassier, Stefan Börno, Bernd Timmermann, Claudia Giesecke-Thiel, Pedro Vallecillo-García, Aris N. Economides, Fabien Le Grand, Petra Knaus, Sven Geissler, Sigmar Stricker
Zbtb14 regulates monocyte and macrophage development through inhibiting pu.1 expression in zebrafish
Yun Deng, Haihong Wang, Xiaohui Liu, Hao Yuan, Jin Xu, Hugues de Thé, Jun Zhou, Jun Zhu
Impact of late larval nutritional stress on adult metabolic, gut and locomotor phenotypes in Drosophila melanogaster
Shri Gouri Patil, Sushmitha Sekhar, Aman Agarwal, TS Oviya, Debashis Rout, Megha
Endothelial Dnmt3a controls placenta vascularization and function to support fetal growth
Stephanie Gehrs, Moritz Jakab, Ewgenija Gutjahr, Zuguang Gu, Dieter Weichenhan, Carolin Mogler, Matthias Schlesner, Christoph Plass, Hellmut G. Augustin, Katharina Schlereth
De-differentiation and Proliferation of Artery Endothelial Cells Drive Coronary Collateral Development
Gauri Arolkar, K. Sneha, Hanjay Wang, Karen M. Gonzalez, Suraj Kumar, Pamela E. Rios Coronado, Y. Joseph Woo, Kristy Red-Horse, Soumyashree Das
CTCF, BEAF-32 and CP190 are not required for the initial establishment of TADs in early Drosophila embryos, but have locus specific roles
Gabriel R. Cavalheiro, Charles Girardot, Rebecca R. Viales, Songjie Feng, Tim Pollex, T. B. Ngoc Cao, Perrine Lacour, Adam Rabinowitz, Eileen E.M. Furlong
Emergent dynamics of adult stem cell lineages from single nucleus and single cell RNA-Seq of Drosophila testes
Amelie A. Raz, Gabriela S. Vida, Sarah R. Stern, Sharvani Mahadevaraju, Jaclyn M. Fingerhut, Jennifer M. Viveiros, Soumitra Pal, Jasmine R. Grey, Mara R. Grace, Cameron W. Berry, Hongjie Li, Jasper Janssens, Wouter Saelens, Zhantao Shao, Chun Hun, Yukiko M. Yamashita, Teresa M. Przytycka, Brian Oliver, Julie A. Brill, Henry M. Krause, Erika L. Matunis, Helen White-Cooper, Stephen DiNardo, Margaret T. Fuller
Inhibition of TGFβ pathway prevents short body size and cardiac defects in Nipbl-deficient mice, a mouse model of Cornelia de Lange syndrome
Céline Hachoud, Faten Chaabani, Erwan Watrin, Valérie Cormier-Daire, Michel Pucéat
Single Cell Multimodal Analyses Reveal Epigenomic and Transcriptomic Basis for Birth Defects in Maternal Diabetes
Tomohiro Nishino, Sanjeev S. Ranade, Angelo Pelonero, Benjamin J. van Soldt, Lin Ye, Michael Alexanian, Frances Koback, Yu Huang, Nandhini Sadagopan, Arun Padmanabhan, Reuben Thomas, Joke G. van Bemmel, Casey A. Gifford, Mauro W. Costa, Deepak Srivastava
Foxm1 drives cardiomyocyte proliferation in adult zebrafish after cardiac injury
Daniel A. Zuppo, Maria A. Missinato, Lucas Santana-Santos, Guang Li, Panayiotis V. Benos, Michael Tsang
Cell cycle and temporal transcription factors regulate proliferation and neuronal diversity of dedifferentiation-derived neural stem cells
Kellie Veen, Francesca Froldi, Qian Dong, Edel Alvarez-Ochoa, Phuong-Khanh Nguyen, Kieran F Harvey, John P D McMullen, Owen Marshall, Patricia R Jusuf, Louise Y Cheng
RET enhancer haplotype-dependent remodeling of the human fetal gut development program
Sumantra Chatterjee, Lauren E. Fries, Or Yaacov, Nan Hu, Hanna E. Berk-Rauch, Aravinda Chakravarti
| Plant development
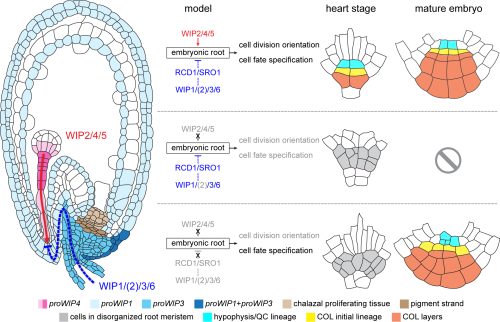
Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium
Riikka Mäkilä, Brecht Wybouw, Ondrej Smetana, Leo Vainio, Anna Solé-Gil, Munan Lyu, Lingling Ye, Xin Wang, Riccardo Siligato, Mark Kubo Jenness, Angus S. Murphy, Ari Pekka Mähönen
The role of GmXTH1, a new xyloglucan endotransglycosylase/hydrolase from soybean, in regulating soybean root growth at seedling stage
Yang Song, Ye Zhang, Ye-yao Du, Sujie Fan, Di Qin, Zhuo Zhang, Pi-wu Wang
Single-cell transcriptomics of the Arabidopsis floral abscission zone
Isaiah W. Taylor, O. Rahul Patharkar, Che-Wei Hsu, John Baer, Chad E. Niederhuth, Uwe Ohler, Philip N. Benfey, John C. Walker
Cortical polarity ensures its own asymmetric inheritance in the stomatal lineage to pattern the leaf surface
Andrew Muroyama, Yan Gong, Kensington S. Hartman, Dominique Bergmann
Exotic alleles of EARLY FLOWERING 3 determine plant development and grain yield in barley
Tanja Zahn, Zihao Zhu, Niklas Ritoff, Jonathan Krapf, Astrid Junker, Thomas Altmann, Thomas Schmutzer, Christian Tüting, Panagiotis L. Kastritis, Marcel Quint, Klaus Pillen, Andreas Maurer
RAV1 mediates cytokinin signalling for regulating primary root growth in Arabidopsis
Drishti Mandal, Saptarshi Datta, Giridhar Ravindra, Pranab Kumar Mondal, Ronita Nag Chaudhuri
High-throughput and automatic structural and developmental root phenotyping on Arabidopsis seedlings
Romain Fernandez, Amandine Crabos, Morgan Maillard, Philippe Nacry, Christophe Pradal
Transcriptional signatures of wheat inflorescence development
Carl VanGessel, James Hamilton, Facundo Tabbita, Jorge Dubcovsky, Stephen Pearce
PAT mRNA decapping factors function specifically and redundantly during development in Arabidopsis
Zhangli Zuo, Milena Edna Roux, Yasin F. Dagdas, Eleazar Rodriguez, Morten Petersen
The circadian clock controls temporal and spatial patterns of floral development in sunflower
Carine M. Marshall, Veronica L. Thompson, Nicky M. Creux, Stacey L. Harmer
Cell surface receptor kinase FERONIA linked to nutrient sensor TORC1 signaling controls root hair growth at low temperature in Arabidopsis thaliana
Javier Martínez Pacheco, Limei Song, Lenka Kuběnová, Miroslav Ovečka, Victoria Berdion Gabarain, Juan Manuel Peralta, Tomás Urzúa Lehuedé, Miguel Angel Ibeas, Sirui Zhu, Yanan Shen, Mikhail Schepetilnikov, Lyubov A Ryabova, José M. Alvarez, Rodrigo A. Gutierrez, Guido Grossman, Jozef Šamaj, Feng Yu, José M. Estevez
REGENERATION VIA SOMATIC EMBRYOGENESIS FROM SEED EXPLANT OF MEDICINAL PLANT SOLANUM VIRGINIANUM (L.)
Dhanashree S. Patil, Swaroopa A. Patil
DEFECTIVELY ORGANIZED TRIBUTARIES 5 is not required for leaf venation patterning in Arabidopsis thaliana
Daniela Vlad, Jane A. Langdale
mRNA decapping machinery targets LBD3/ASL9 transcripts to allow developmental changes in Arabidopsis
Zhangli Zuo, Milena Edna Roux, Jonathan Renaud Chevalier, Yasin F. Dagdas, Takafumi Yamashino, Søren Diers Højgaard, Emilie Knight, Lars Østergaard, Eleazar Rodriguez, Morten Petersen
Seed reserve mobilization and seedling morphology in a bioassay for the detection of genetically modified soybean
Francisco Cleilson Lopes Costa, Samanda López Peña, Welison Andrade Pereira
| Evo-devo
The effect of developmental pleiotropy on the evolution of insect immune genes
Thi Minh Ngo, Alissa M. Williams, Ann T. Tate
Growth rate as a modulator of tooth patterning during adaptive radiations
Alexa Sadier, Neal Anthwal, Andrew L. Krause, Renaud Dessalles, Michael Lake, Laurent Bentolila, Robert Haase, Natalie Nieves, Sharlene Santana, Karen Sears
The interplay between developmental stage and environment underlies the adaptive effect of a natural transposable element insertion
Miriam Merenciano, Josefa González
Clonal development, not aggregation, drives the transition to multicellularity in an isogenic model system
Jennifer T. Pentz, Kathryn MacGillivray, James G. DuBose, Peter L. Conlin, Emma Reinhardt, Eric Libby, William C. Ratcliff
Teeth outside the mouth: the evolution and development of shark denticles
Rory L. Cooper, Ella F. Nicklin, Liam J. Rasch, Gareth J. Fraser
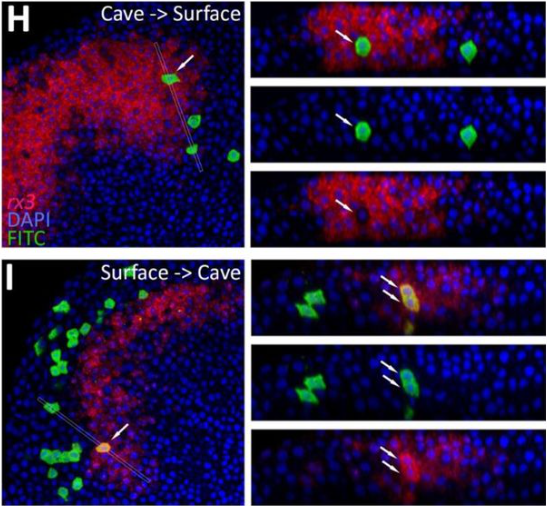
Evolution of the regulation of developmental gene expression in blind Mexican cavefish
Julien Leclercq, Jorge Torres-Paz, Maxime Policarpo, François Agnès, Sylvie Rétaux
Origins of smooth muscle and evolutionary specializations of the pulmonary mesenchyme in the vertebrate lung
Katharine Goodwin, Michael A. Palmer, Bezia Lemma, Celeste M. Nelson
Brachiopod and mollusc biomineralisation is a conserved process that was lost in the phoronid-bryozoan stem lineage
Joel Vikberg Wernström, Ludwik Gąsiorowski, Andreas Hejnol
Phenotypic plasticity, life cycles, and the evolutionary transition to multicellularity
Si Tang, Yuriy Pichugin, Katrin Hammerschmidt
The mammalian forelimb diversity as a morphological gradient of increasing evolutionary versatility
Priscila S. Rothier, Anne-Claire Fabre, Julien Clavel, Roger Benson, Anthony Herrel
Cell Biology
Kindlin-2 inhibits TNF/NF-κB-caspase 8 pathway in hepatocytes to maintain liver development and function
Huanqing Gao, Yiming Zhong, Liang Zhou, Sixiong Lin, Xiaoting Hou, Zhen Ding, Yan Li, Qing Yao, Huiling Cao, Xuenong Zou, Di Chen, Xiaochun Bai, Guozhi Xiao
A CDKB/KRP/FB3 cell cycle core complex functions in rice gametes and zygotes
Hengping Xu, Laura Bartley, Marc Libault, Venkatesan Sundaresan, Hong Fu, Scott Russell
Acquisition of the Spindle Assembly Checkpoint and its modulation by cell fate and cell size in a chordate embryo
Marianne Roca, Lydia Besnardeau, Elisabeth Christians, Alex McDougall, Janet Chenevert, Stefania Castagnetti
Meiotic and mitotic aneuploidies drive arrest of in vitro fertilized human preimplantation embryos
Rajiv C. McCoy, Michael C. Summers, Abeo McCollin, Christian S. Ottolini, Kamal Ahuja, Alan H. Handyside
Annexin A1 is a polarity cue that directs planar mitotic spindle orientation during mammalian epithelial morphogenesis
Maria Fankhaenel, Farahnaz Sadat Golestan Hashemi, Larissa Mourao, Emily Lucas, Manal Mosa Hosawi, Paul Skipp, Xavier Morin, Colinda L.G.J. Scheele, Salah Elias
Fluorescence spectroscopy of low-level endogenous β-adrenergic receptor expression at the plasma membrane of differentiating human iPSC-derived cardiomyocytes
Philipp Gmach, Marc Bathe-Peters, Narasimha Telugu, Martin J Lohse, Paolo Annibale
Pseudouridine-dependent ribosome biogenesis regulates translation of polyglutamine proteins during Drosophila oogenesis
Shane Breznak, Yingshi Peng, Limin Deng, Noor M. Kotb, Zachary Flamholz, Ian T. Rapisarda, Elliot T. Martin, Kara A. LaBarge, Dan Fabris, Elizabeth R. Gavis, Prashanth Rangan
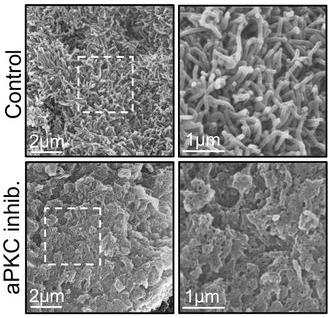
Loss of cell polarity regulators initiates pyroptosis in trophoblasts at the human maternal fetal interface
Khushali Patel, Jasmine Nguyen, Sumaiyah Shaha, Ashley Zubkowski, Meghan Riddell
GJA1 Depletion Causes Ciliary Defects by Affecting Rab11 Trafficking to the Ciliary Base
Dong Gil Jang, Keun Yeong Kwon, Yeong Cheon Kweon, Byung-gyu Kim, Kyungjae Myung, Hyun-Shik Lee, Chan Young Park, Taejoon Kwon, Tae Joo Park
The serine/threonine kinase Back seat driver prevents cell fusion to maintain cell identity
Shuo Yang, Aaron N. Johnson
Propagation dynamics of electrotactic motility in large epithelial cell sheets
Yan Zhang, Guoqing Xu, Jiandong Wu, Rachel M Lee, Zijie Zhu, Yaohui Sun, Kan Zhu, Wolfgang Losert, Simon Liao, Gong Zhang, Tingrui Pan, Zhengping Xu, Francis Lin, Min Zhao
Dynamic states of cervical epithelia during pregnancy and epithelial barrier disruption
Anne Cooley, ShanmugaPriyaa Madhukaran, Elizabeth Stroebele, Mariano Colon Caraballo, Lei Wang, Gary C. Hon, Mala Mahendroo
Modelling
Model of neural induction in the ascidian embryo
Rossana Bettoni, Clare Hudson, Hitoyoshi Yasuo, Sophie de Buyl, Geneviève Dupont
Competency of the Developmental Layer Alters Evolutionary Dynamics in an Artificial Embryogeny Model of Morphogenesis
Lakshwin Shreesha, Michael Levin
A mathematical modelling portrait of Wnt signalling in early vertebrate embryogenesis
Claudiu V. Giuraniuc, Shabana Zain, Shahmama Ghafoor, Stefan Hoppler
Is cell segregation like oil and water: asymptotic versus transitory regime
Florian Franke, Sebatian Aland, Hans-Joachim Böhme, Anja Voss-Böhme, Steffen Lange
Reviews
The History, Current Status, Benefits, and Challenges of 3D Printed Organs
Alicia Shin, Sumin Kim
Mechanics of Morphogenesis in Neural Development: in vivo, in vitro, and in silico
Joseph Sutlive, Hamed Seyyedhosseinzadeh, Zheng Ao, Haning Xiu, Kun Gou, Feng Guo, Zi Chen
SUMOylation in Skeletal Development, Homeostasis, and Disease
Tao Yang , Huadie Liu , Sonya E. L. Craig , Vladimir Molchanov , Joe Floramo , Yaguang Zhao
Tools & Resources
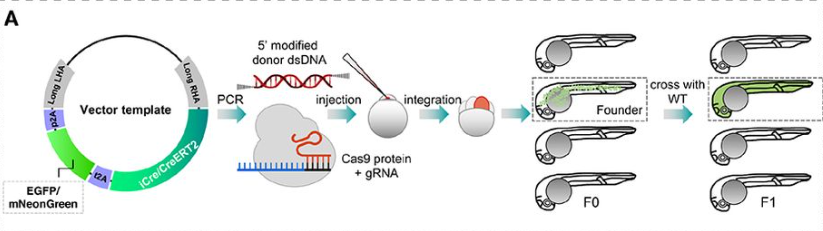
Efficient knock-in method enabling lineage tracing in zebrafish
Jiarui Mi, Olov Andersson
Accurate simultaneous sequencing of genetic and epigenetic bases in DNA
Jens Füllgrabe, Walraj S Gosal, Páidí Creed, Sidong Liu, Casper K Lumby, David J Morley, Tobias W B Ost, Albert J Vilella, Shirong Yu, Helen Bignell, Philippa Burns, Tom Charlesworth, Beiyuan Fu, Howerd Fordham, Nick Harding, Olga Gandelman, Paula Golder, Christopher Hodson, Mengjie Li, Marjana Lila, Yang Liu, Joanne Mason, Jason Mellad, Jack Monahan, Oliver Nentwich, Alexandra Palmer, Michael Steward, Minna Taipale, Audrey Vandomme, Rita Santo San-Bento, Ankita Singhal, Julia Vivian, Natalia Wójtowicz, Nathan Williams, Nicolas J Walker, Nicola C H Wong, Gary Yalloway, Joanna D Holbrook, Shankar Balasubaramanian
CeDAR: incorporating cell type hierarchy improves cell type specific differential analyses in bulk omics data
Luxiao Chen, Ziyi Li, Hao Wu
Retaining pluripotency and exogenous mRNA introduction in planarian stem cell culture
Kai Lei, Wenya Zhang, Jiajia Chen, Sean A. McKinney, Eric J. Ross, Heng-Chi Lee, Alejandro Sánchez Alvarado
Maximizing CRISPRi efficacy and accessibility with dual-sgRNA libraries and optimal effectors
Joseph M. Replogle, Jessica L. Bonnar, Angela N. Pogson, Christina R. Liem, Nolan K. Maier, Yufang Ding, Baylee J. Russell, Xingren Wang, Kun Leng, Alina Guna, Thomas M. Norman, Ryan A. Pak, Daniel M. Ramos, Michael E. Ward, Luke A. Gilbert, Martin Kampmann, Jonathan S. Weissman, Marco Jost
Establishment and Characterization of Novel Canine Organoids with Organ-Specific Physiological Similarity
Christopher Zdyrski, Vojtech Gabriel, Oscar Ospina, Hannah Wickham, Dipak K. Sahoo, Kimberly Dao, Leeann S. Aguilar Meza, Leila Bedos, Sydney Honold, Pablo Piñeyro, Jonathan P. Mochel, Karin Allenspach
DrosOmics: the comparative genomics browser to explore omics data in natural strains of D. melanogaster
Marta Coronado-Zamora, Judit Salces-Ortiz, Josefa González
SMAP design: A multiplex PCR amplicon and gRNA design tool to screen for natural and CRISPR-induced genetic variation
Ward Develtere, Evelien Waegneer, Kevin Debray, Sabine Van Glabeke, Steven Maere, Tom Ruttink, Thomas B. Jacobs
The presence of BBB hastens neuronal differentiation of cerebral organoids – the potential role of endothelial derived BDNF
Giorgia Fedele, Alessandra Cazzaniga, Sara Castiglioni, Laura Locatelli, Antonella Tosoni, Manuela Nebuloni, Jeanette A. M. Maier
PhenoTrack3D: an automatic high-throughput phenotyping pipeline to track maize organs over time
Benoit Daviet, Romain Fernandez, Llorenç Cabrera-Bosquet, Christophe Pradal, Christian Fournier
Molecular characterization of the intact muscle spindle using a multi-omics approach
Bavat Bornstein, Lia Heinemann-Yerushalmi, Sharon Krief, Ruth Adler, Bareket Dassa, Dena Leshkowitz, Minchul Kim, Guy Bewick, Robert W. Banks, Elazar Zelzer
Generation of a transparent killifish line through multiplex CRISPR/Cas9-mediated gene inactivation
Johannes Krug, Carolin Albertz, Vera L. Hopfenmüller, Christoph Englert
Neuro-mesodermal assembloids recapitulate aspects of peripheral nervous system development in vitro
Anna F. Rockel, Nicole Wagner, Süleyman Ergün, Philipp Wörsdörfer
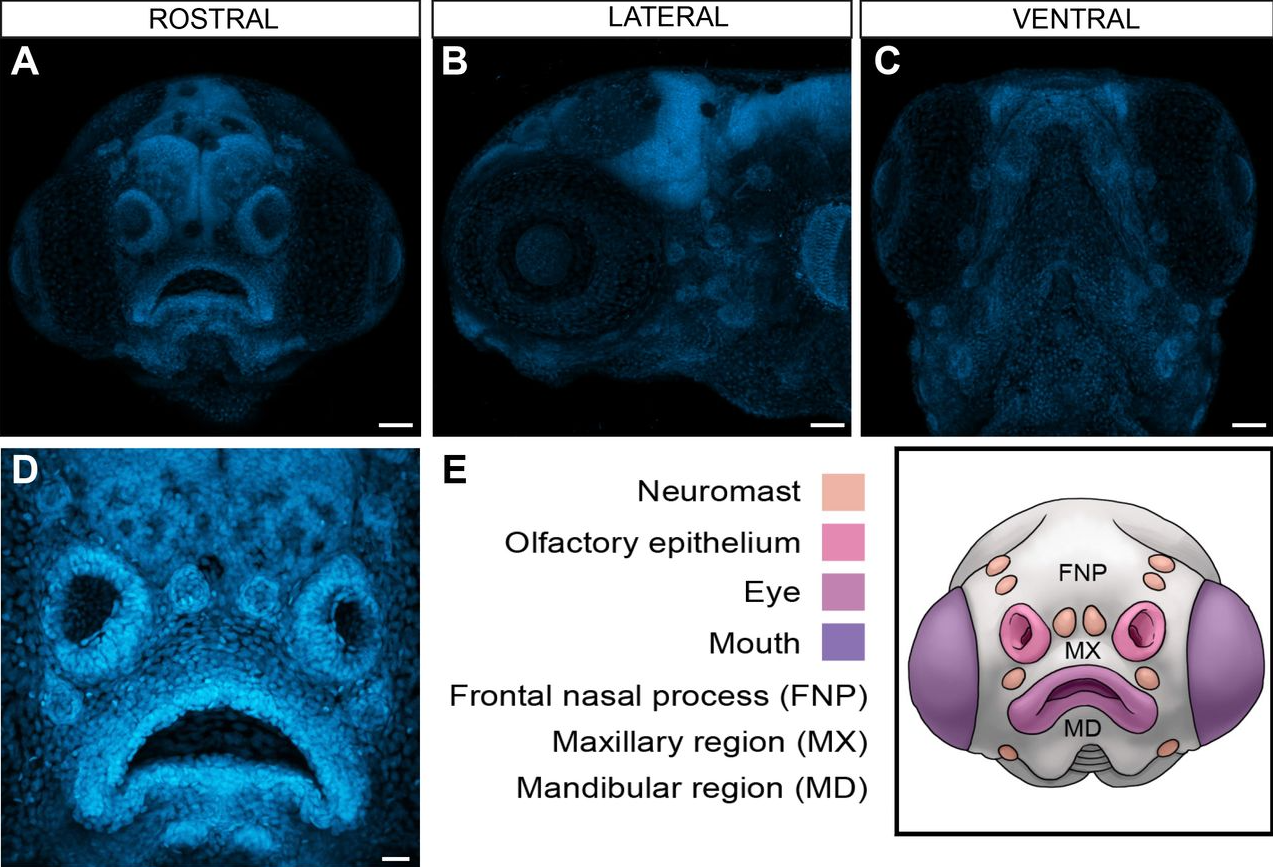
zFACE: Facial Analytics from a Coordinate Extrapolation System for Developing Zebrafish
Lorena Maili, Oscar E. Ruiz, Philip Kahan, Stephen T. Larson, S. Shahrukh Hashmi, Jacqueline T. Hecht, George T. Eisenhoffer
Efficient generation of marmoset primordial germ cell-like cells using induced pluripotent stem cells
Yasunari Seita, Keren Cheng, John R. McCarrey, Nomesh Yadu, Ian Cheeseman, Alec Bagwell, Corinna N. Ross, Isamar Santana-Toro, Li-Hua Yen, Sean Vargas, Christopher S. Navara, Brian P. Hermann, Kotaro Sasaki
Research practice & education
RMeDPower for Biology: guiding design, experimental structure and analyses of repeated measures data for biological studies
Min-Gyoung Shin, Julia A. Kaye, Naufa Amirani, Stephanie Lam, Reuben Thomas, Steven Finkbeiner
If this title is funny, will you cite me? Citation impacts of humour and other features of article titles in ecology and evolution
Stephen B. Heard, Chloe A. Cull, Easton R. White


 (No Ratings Yet)
(No Ratings Yet)

 (2 votes)
(2 votes)