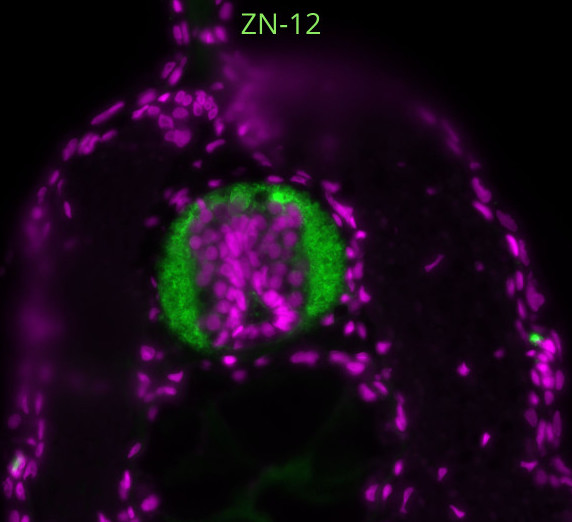
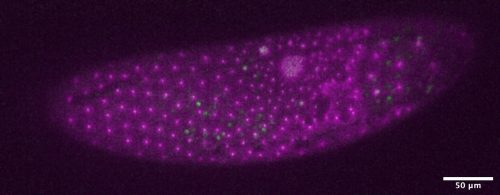
In skeletal muscle, we know that two stem/progenitors, muscle satellite cells (MuSCs) and mesenchymal progenitors (also known as FAPs), are critical for homeostasis, regeneration, and growth including muscle hypertrophy. Although the involvement of MuSCs in muscle hypertrophy has been well studied, the mechanism underling MuSC proliferation had not been investigated. One reason for this omission was that MuSC proliferation was thought to occur by the same mechanism as the well-studied process of muscle regeneration and muscle loading. In addition, the roles of mesenchymal progenitors in muscle hypertrophy had not been elucidated. Based on this background, we started this project and recently published results (Kaneshige et al., Cell Stem Cell, 2022) that shed light on the mechanisms regulating MuSC proliferation by mesenchymal progenitors in a surgical loaded muscle. In this paper, we presented data that showed 1) critical roles of mesenchymal progenitors in MuSC proliferation, 2) involvement of Yap1/Taz in loaded mesenchymal progenitors, 3) the contribution of mesenchymal progenitors-derived Thrombospondin-1 (Thbs1) as the target of Yap1/Taz to MuSC proliferation, and 4) Thbs1-derived peptide induces the proliferation of Calcitonin receptor (CalcR)-mutant MuSCs. However, the results were not obtained in the order shown in the figures; for example, the analysis of Pa-Yap1/Taz was the last experiment of the main project. In this blog, the results will be presented in the order in which they were discovered.
Depletion of mesenchymal progenitors in loaded muscle
In our previous project (Fukuda et al., 2019), we had noticed morphological changes in mesenchymal progenitors by mechanical load using FACS analyses (actually, we did not put a high priority on it due to our focusing on MuSCs at that time…). These data and our new hypothesis, that mesenchymal progenitors might be involved in muscle hypertrophy, encouraged us to initiate this project. Fortunately, our team (Professor Uezumi, co-corresponding author) had already established an experimental model for depleting mesenchymal progenitors. We started to collaborate before his first publication using Pa-DTA (PdgfraCreERT::Rosa DTA) mice (Uezumi et al., 2021). We were very excited when our data convinced us that the expansion of MuSCs during overload was significantly reduced in Pa-DTA mice depleted mesenchymal progenitors (Kaneshige et al., 2021).
Microarray analysis of mesenchymal progenitors
How do mesenchymal progenitors regulate MuSCs in loaded muscles? To address this crucial question, we conducted microarray analysis of mesenchymal progenitors from loaded muscle and explored MuSC regulators. We focused on genes with the following three features; 1) upregulated by mechanical loading, 2) secreted molecules, 3) different expression pattern from muscle injury model. We finally got four candidates including Thbs1. Since the gene expression analysis of Pa-Yap1/Taz-cdKO, which will be described later, was performed by RNA-seq, we re-analyzed normal mesenchymal progenitors by RNA-seq for consistency.
Screening of MuSCs regulator candidates
To uncover a signaling mechanism between mesenchymal progenitors and MuSCs, we conducted an in vivo screening with inhibitory antibody against the above candidates or their receptor. For Thbs1, we tested an antibody against CD47, which is one of well-known receptor of Thbs1 and highly expressed in MuSCs. As all antibodies were commercially available, this was a feasible strategy. Thus, we analyzed the loaded muscles and found that new myonuclei supply was reduced in mice treated with anti-CD47 antibody.
Injection of PKHB1 into CalcR-cKO
Elucidation of the role of CalcR in MuSCs has been one of the main themes in our laboratory (Baghdadi et al., 2018; Fukada et al., 2007; Yamaguchi et al., 2015; Zhang et al., 2019). CalcR-mutated MuSCs exhibit the increased expression of cell cycle-related genes including Ki67. However, no substantial cell division was observed. In contrast, MuSC number was decreased in time dependent manner after CalcR-depletion by tamoxifen injection.
Early on this project, we had observed that CalcR expression in MuSCs were decreased in the surgical loaded muscle, however, no further analysis had been conducted. After that, we found that PKHB1 (CD47 agonist peptide designed based on CD47 binding sequence of Thbs1) promoted MuSC proliferation in loaded muscle, but not in non-loaded muscle. To know the effect of CalcR on MuSC expansion by PKHB1, we tried to inject PKHB1 into sedentary CalcR-cKO mice without surgical muscle overload. In these analyses, MuSC number on isolated myofibers from the treated mice were counted in blind manner. We were very surprised to see that the number of MuSCs in CalcR-cKO was increased by PKHB1 injection. We also observed the MuSCs-derived new myonuclei which were rarely detected in untreated CalcR-cKO mice.
Yap1/Taz in mesenchymal progenitors
As the downstream target of CalcR signaling, we had previously focused on Yap1 (Zhang et al., 2019), therefore, we already had Yap1-floxed mice. This meant that it was easy for us to generate PdgfraCreER::Yap1flox/flox mice. As Yap1 is known as a mechanical transducer, we expected that the surgical loaded model would be nice way to know the role of Yap1 in mesenchymal progenitors. However, we could not detect consistent difference between control and Pa-Yap1 (PdgfraCreERT::Yap1flox/flox). Therefore, by collaborating with Professor Potent, Dr Watanabe, and Professor Braun, we generated Pa-Y/T-cdKO (PdgfraCreERT::Yap1flox/flox::Tazflox/flox) mice, and could observe the remarkable differences between control and Pa-Y/T-cdKO mice.
RNA-seq analyses links Yap1/Taz and Thbs1
As mentioned above, we had already obtained the data indicating that the blockage of CD47 inhibited the increased number of MuSCs-derived myonuclei and that the CD47 agonist, PKHB1, promoted MuSCs proliferation in vivo. As CD47 expression is ubiquitously expressed in many types of cells, we needed to use MuSCs-specific CD47-depletion mice. Fortunately, Professor Matoba and Dr Saito kindly provided us CD47-floxed mice, which enabled us to analyze MuSC-specific CD47-cKO (Pax7CreERT2::Cd47 flox/flox) mice.
In collaboration with Professor Ohkawa and Dr Maehara, we performed RNA-seq analysis of Pa-Y/T-cdKO mesenchymal progenitors and our data demonstrated reduced expression of Thbs1 as well as Yap-target genes. By performing some additional experiments including nice immunostaining results from Professor Uezumi’s team, we were sure that Thbs1 is one critical target by Yap1/Taz in loaded mesenchymal progenitors and promotes MuSCs proliferation via CD47.
In the end, we are really grateful to all authors involved in this project. Sometimes, the research object already exists closed to you, or sometimes, initial data has already been in your hand, which might be serendipity. We already had HeyL-deficient mice which provided the basis of this project (Fukuda et al., 2019). Kobe University, where CD47-floxed mice were housed in Professor Matoda Lab., is geographically very close to Osaka University. This project taught us that critical materials or animals could already be close to us and that new collaborations can provide unexpected links between different projects.
References
Baghdadi, M.B., Castel, D., Machado, L., Fukada, S.I., Birk, D.E., Relaix, F., Tajbakhsh, S., and Mourikis, P. (2018). Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature 557, 714-718.
Fukada, S., Uezumi, A., Ikemoto, M., Masuda, S., Segawa, M., Tanimura, N., Yamamoto, H., Miyagoe-Suzuki, Y., and Takeda, S. (2007). Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 25, 2448-2459.
Fukuda, S., Kaneshige, A., Kaji, T., Noguchi, Y.T., Takemoto, Y., Zhang, L., Tsujikawa, K., Kokubo, H., Uezumi, A., Maehara, K., et al. (2019). Sustained expression of HeyL is critical for the proliferation of muscle stem cells in overloaded muscle. Elife 8, e48284.
Kaneshige, A., Kaji, T., Zhang, L., Saito, H., Nakamura, A., Kurosawa, T., Ikemoto-Uezumi, M., Tsujikawa, K., Seno, S., Hori, M., et al. (2022). Relayed signaling between mesenchymal progenitors and muscle stem cells ensures adaptive stem cell response to increased mechanical load. Cell Stem Cell 29, 265-280 e266.
Uezumi, A., Ikemoto-Uezumi, M., Zhou, H., Kurosawa, T., Yoshimoto, Y., Nakatani, M., Hitachi, K., Yamaguchi, H., Wakatsuki, S., Araki, T., et al. (2021). Mesenchymal Bmp3b expression maintains skeletal muscle integrity and decreases in age-related sarcopenia. J Clin Invest 131, e139617.
Yamaguchi, M., Watanabe, Y., Ohtani, T., Uezumi, A., Mikami, N., Nakamura, M., Sato, T., Ikawa, M., Hoshino, M., Tsuchida, K., et al. (2015). Calcitonin Receptor Signaling Inhibits Muscle Stem Cells from Escaping the Quiescent State and the Niche. Cell reports 13, 302-314.
Zhang, L., Noguchi, Y.T., Nakayama, H., Kaji, T., Tsujikawa, K., Ikemoto-Uezumi, M., Uezumi, A., Okada, Y., Doi, T., Watanabe, S., et al. (2019). The CalcR-PKA-Yap1 Axis Is Critical for Maintaining Quiescence in Muscle Stem Cells. Cell reports 29, 2154-2163 e2155.
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)

 (1 votes)
(1 votes)