Frontiers in Stem Cell and Organoid Medicine Symposium
Posted by Helen Zenner, on 11 February 2022
For more information and to register: https://www.organoidmedicinesymposium.org/

Posted by Helen Zenner, on 11 February 2022
For more information and to register: https://www.organoidmedicinesymposium.org/

Posted by the Node, on 10 February 2022
One of the key objectives of the Node Network is to allow scientists, especially early-career researchers (ECRs), to raise their profiles in the developmental and stem cell biology community. With this in mind, and as part of our second birthday celebrations, we are delighted to launch our discussion and networking event, ‘Promoting yourself as an ECR’ hosted with our sister community sites FocalPlane and preLights. The interactive event will begin with a panel discussion with our three invited panellists Maria Abou Chakra, Pablo Sáez and Sarvenaz Sarabipour and then continue with a networking event where you can meet the panellists, representatives from The Company of Biologists and other ECRs.

Dr Maria Abou Chakra is a research associate at the University of Toronto, where her research focuses on mathematical modelling of stem cell development. She is the organiser of the Modelling Cell Development & Regeneration Discussion Group, as well as having been involved in outreach, mentorship and EDI events.
Professor Pablo J. Sáez is a new PI at UKE, Hamburg, where his international team is studying the role of cell communication and migration, with a particular focus on immune cells. Pablo is a regular contributor to our preprint posts on FocalPlane, and is an advocate for better representation for ECRs in academic conferences.
Dr Sarvenaz Sarabipour is an assistant research scientist at Johns Hopkins University, where her research focuses on receptor signalling networks at the cell and tissue level. As well as her scientific interests, she is an active advocate for ECRs, open science, mentorship, and diversity in science.
Posted by Kat Arney, on 10 February 2022
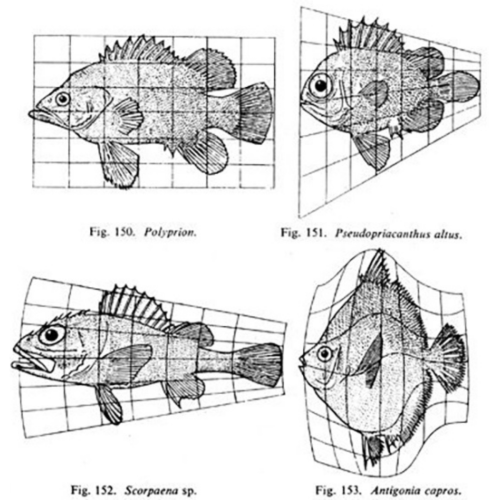
Whereas previously, most biologists felt that the living world had to be dealt with in a completely separate way from the non-living world, D’Arcy was saying, “Actually, no. We can look at a lot of basic forms in biology and explain their growth and shapes according to fundamental laws of physics and mathematics. A living thing may grow in exactly the same way as a non-living thing because it’s just obeying these fundamental laws.”
Matthew Jarron describing D’Arcy Thompson
In the latest episode of the Genetics Unzipped podcast, we’re exploring the life and work of D’Arcy Wentworth Thompson – one of the first scientists to bring together the worlds of mathematics and biology in the quest to understand how living things are built. Dr Kat Arney sat down with the curator of the University of Dundee’s D’Arcy Thompson Zoology Museum, Matthew Jarron, to find out more about this larger than life character…
Genetics Unzipped is the podcast from The Genetics Society. Full transcript, links and references available online at GeneticsUnzipped.com.
Subscribe from Apple podcasts, Spotify, or wherever you get your podcasts.
Head over to GeneticsUnzipped.com to catch up on our extensive back catalogue.
If you enjoy the show, please do rate and review on Apple podcasts and help to spread the word on social media. And you can always send feedback and suggestions for future episodes and guests to podcast@geneticsunzipped.com Follow us on Twitter – @geneticsunzip
Posted by Akihiro Kaneshige, on 9 February 2022
In skeletal muscle, we know that two stem/progenitors, muscle satellite cells (MuSCs) and mesenchymal progenitors (also known as FAPs), are critical for homeostasis, regeneration, and growth including muscle hypertrophy. Although the involvement of MuSCs in muscle hypertrophy has been well studied, the mechanism underling MuSC proliferation had not been investigated. One reason for this omission was that MuSC proliferation was thought to occur by the same mechanism as the well-studied process of muscle regeneration and muscle loading. In addition, the roles of mesenchymal progenitors in muscle hypertrophy had not been elucidated. Based on this background, we started this project and recently published results (Kaneshige et al., Cell Stem Cell, 2022) that shed light on the mechanisms regulating MuSC proliferation by mesenchymal progenitors in a surgical loaded muscle. In this paper, we presented data that showed 1) critical roles of mesenchymal progenitors in MuSC proliferation, 2) involvement of Yap1/Taz in loaded mesenchymal progenitors, 3) the contribution of mesenchymal progenitors-derived Thrombospondin-1 (Thbs1) as the target of Yap1/Taz to MuSC proliferation, and 4) Thbs1-derived peptide induces the proliferation of Calcitonin receptor (CalcR)-mutant MuSCs. However, the results were not obtained in the order shown in the figures; for example, the analysis of Pa-Yap1/Taz was the last experiment of the main project. In this blog, the results will be presented in the order in which they were discovered.
In our previous project (Fukuda et al., 2019), we had noticed morphological changes in mesenchymal progenitors by mechanical load using FACS analyses (actually, we did not put a high priority on it due to our focusing on MuSCs at that time…). These data and our new hypothesis, that mesenchymal progenitors might be involved in muscle hypertrophy, encouraged us to initiate this project. Fortunately, our team (Professor Uezumi, co-corresponding author) had already established an experimental model for depleting mesenchymal progenitors. We started to collaborate before his first publication using Pa-DTA (PdgfraCreERT::Rosa DTA) mice (Uezumi et al., 2021). We were very excited when our data convinced us that the expansion of MuSCs during overload was significantly reduced in Pa-DTA mice depleted mesenchymal progenitors (Kaneshige et al., 2021).
How do mesenchymal progenitors regulate MuSCs in loaded muscles? To address this crucial question, we conducted microarray analysis of mesenchymal progenitors from loaded muscle and explored MuSC regulators. We focused on genes with the following three features; 1) upregulated by mechanical loading, 2) secreted molecules, 3) different expression pattern from muscle injury model. We finally got four candidates including Thbs1. Since the gene expression analysis of Pa-Yap1/Taz-cdKO, which will be described later, was performed by RNA-seq, we re-analyzed normal mesenchymal progenitors by RNA-seq for consistency.
To uncover a signaling mechanism between mesenchymal progenitors and MuSCs, we conducted an in vivo screening with inhibitory antibody against the above candidates or their receptor. For Thbs1, we tested an antibody against CD47, which is one of well-known receptor of Thbs1 and highly expressed in MuSCs. As all antibodies were commercially available, this was a feasible strategy. Thus, we analyzed the loaded muscles and found that new myonuclei supply was reduced in mice treated with anti-CD47 antibody.
Elucidation of the role of CalcR in MuSCs has been one of the main themes in our laboratory (Baghdadi et al., 2018; Fukada et al., 2007; Yamaguchi et al., 2015; Zhang et al., 2019). CalcR-mutated MuSCs exhibit the increased expression of cell cycle-related genes including Ki67. However, no substantial cell division was observed. In contrast, MuSC number was decreased in time dependent manner after CalcR-depletion by tamoxifen injection.
Early on this project, we had observed that CalcR expression in MuSCs were decreased in the surgical loaded muscle, however, no further analysis had been conducted. After that, we found that PKHB1 (CD47 agonist peptide designed based on CD47 binding sequence of Thbs1) promoted MuSC proliferation in loaded muscle, but not in non-loaded muscle. To know the effect of CalcR on MuSC expansion by PKHB1, we tried to inject PKHB1 into sedentary CalcR-cKO mice without surgical muscle overload. In these analyses, MuSC number on isolated myofibers from the treated mice were counted in blind manner. We were very surprised to see that the number of MuSCs in CalcR-cKO was increased by PKHB1 injection. We also observed the MuSCs-derived new myonuclei which were rarely detected in untreated CalcR-cKO mice.
As the downstream target of CalcR signaling, we had previously focused on Yap1 (Zhang et al., 2019), therefore, we already had Yap1-floxed mice. This meant that it was easy for us to generate PdgfraCreER::Yap1flox/flox mice. As Yap1 is known as a mechanical transducer, we expected that the surgical loaded model would be nice way to know the role of Yap1 in mesenchymal progenitors. However, we could not detect consistent difference between control and Pa-Yap1 (PdgfraCreERT::Yap1flox/flox). Therefore, by collaborating with Professor Potent, Dr Watanabe, and Professor Braun, we generated Pa-Y/T-cdKO (PdgfraCreERT::Yap1flox/flox::Tazflox/flox) mice, and could observe the remarkable differences between control and Pa-Y/T-cdKO mice.
As mentioned above, we had already obtained the data indicating that the blockage of CD47 inhibited the increased number of MuSCs-derived myonuclei and that the CD47 agonist, PKHB1, promoted MuSCs proliferation in vivo. As CD47 expression is ubiquitously expressed in many types of cells, we needed to use MuSCs-specific CD47-depletion mice. Fortunately, Professor Matoba and Dr Saito kindly provided us CD47-floxed mice, which enabled us to analyze MuSC-specific CD47-cKO (Pax7CreERT2::Cd47 flox/flox) mice.
In collaboration with Professor Ohkawa and Dr Maehara, we performed RNA-seq analysis of Pa-Y/T-cdKO mesenchymal progenitors and our data demonstrated reduced expression of Thbs1 as well as Yap-target genes. By performing some additional experiments including nice immunostaining results from Professor Uezumi’s team, we were sure that Thbs1 is one critical target by Yap1/Taz in loaded mesenchymal progenitors and promotes MuSCs proliferation via CD47.
In the end, we are really grateful to all authors involved in this project. Sometimes, the research object already exists closed to you, or sometimes, initial data has already been in your hand, which might be serendipity. We already had HeyL-deficient mice which provided the basis of this project (Fukuda et al., 2019). Kobe University, where CD47-floxed mice were housed in Professor Matoda Lab., is geographically very close to Osaka University. This project taught us that critical materials or animals could already be close to us and that new collaborations can provide unexpected links between different projects.

Baghdadi, M.B., Castel, D., Machado, L., Fukada, S.I., Birk, D.E., Relaix, F., Tajbakhsh, S., and Mourikis, P. (2018). Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature 557, 714-718.
Fukada, S., Uezumi, A., Ikemoto, M., Masuda, S., Segawa, M., Tanimura, N., Yamamoto, H., Miyagoe-Suzuki, Y., and Takeda, S. (2007). Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 25, 2448-2459.
Fukuda, S., Kaneshige, A., Kaji, T., Noguchi, Y.T., Takemoto, Y., Zhang, L., Tsujikawa, K., Kokubo, H., Uezumi, A., Maehara, K., et al. (2019). Sustained expression of HeyL is critical for the proliferation of muscle stem cells in overloaded muscle. Elife 8, e48284.
Kaneshige, A., Kaji, T., Zhang, L., Saito, H., Nakamura, A., Kurosawa, T., Ikemoto-Uezumi, M., Tsujikawa, K., Seno, S., Hori, M., et al. (2022). Relayed signaling between mesenchymal progenitors and muscle stem cells ensures adaptive stem cell response to increased mechanical load. Cell Stem Cell 29, 265-280 e266.
Uezumi, A., Ikemoto-Uezumi, M., Zhou, H., Kurosawa, T., Yoshimoto, Y., Nakatani, M., Hitachi, K., Yamaguchi, H., Wakatsuki, S., Araki, T., et al. (2021). Mesenchymal Bmp3b expression maintains skeletal muscle integrity and decreases in age-related sarcopenia. J Clin Invest 131, e139617.
Yamaguchi, M., Watanabe, Y., Ohtani, T., Uezumi, A., Mikami, N., Nakamura, M., Sato, T., Ikawa, M., Hoshino, M., Tsuchida, K., et al. (2015). Calcitonin Receptor Signaling Inhibits Muscle Stem Cells from Escaping the Quiescent State and the Niche. Cell reports 13, 302-314.
Zhang, L., Noguchi, Y.T., Nakayama, H., Kaji, T., Tsujikawa, K., Ikemoto-Uezumi, M., Uezumi, A., Okada, Y., Doi, T., Watanabe, S., et al. (2019). The CalcR-PKA-Yap1 Axis Is Critical for Maintaining Quiescence in Muscle Stem Cells. Cell reports 29, 2154-2163 e2155.
Posted by Arif Ashraf, on 8 February 2022
Plant Postdocs started the journey as an organization in September 2019, a few months before the pandemic. At the beginning, our goal was to organize virtual webinars, seminars, and career sessions, where postdocs could share their works and sessions about job opportunities and professional development. As a small start-up organization with leadership team members from different institutions, internet was the only way for us to reach our peers. But, back in our mind we knew that it is challenging to organize and run every event online. People are not very used to the idea of virtual events, and most importantly not properly equipped and comfortable.
At the beginning of the 2020, the COVID-19 spread forced universities and institutes to go online and cancel in-person classes and seminars. Even before the lockdown was announced in February, we scheduled our first two virtual seminars. By March 2020, when we had our first webinar, serendipitously it was the first few weeks of pandemic lockdown. To our surprise, the timing for a virtual webinar could not be better than that. That was the beginning of the virtual webinar series of Plant Postdocs. We had a really good participation and feedback from the community, but we also realized there is room to improve, a better way to reach our peers, and engage the community.

Besides the events organized in each month or semester, we created the day to day communication platform on Slack. Slack is very organized to keep our conversation focused in different channels. Like any other virtual platform, Slack took time to become a popular way of communication among scientific society. Plant Postdocs’ experience is no different in this case. Compared to our official Twitter platform (which has more than 5k followers), we have currently 300+ members on Slack. We post our activities on both platforms, Twitter and Slack; and we do not want anyone to miss important events and updates just because they are not present or active in one of those platforms. In addition, we have an autobot in our Slack as a channel, where all of our tweets appear automatically. Although the group is smaller in Slack, so far this is the best way to do the community conversation and pin the link of important resources as well.
Since the beginning to recent days, unfortunately we are not completely out of COVID-19 era. This practically means no in-person conferences for two academic years. However, the Plant Postdocs’ webinars were an opportunity for postdocs to present their work when in-person conferences were canceled. The cancellation of in-person events led people to adapt better for the virtual events. Over the time, we have observed that panelists and participants are more engaged and comfortable in virtual events. Even as a regular organizer, we find it easier to organize events these days compared to the beginning.
Virtual events helped us to keep going through the difficult pandemic lockdown time. At the same time, everyone’s schedule was full of multiple Zoom events in a day. Too many virtual events caused “Zoom fatigue” in the meantime. As a matter of fact, most virtual events started to observe less and less participants over the time. Plant Postdocs’ events were no exception in this case. To overcome the declining participants, we have decided to diversify our webinar topics.
Our organization is dedicated to postdocs in the plant biology field. At the beginning, we used to organize two career webinars, one about academic jobs and another for industry/Government jobs, in each semester. Along with Zoom fatigue, highlighting similar topics in the webinars again and again made it monotonous to our regular participants. To overcome this issue, we organized webinars on scientific writing and editing, preparation for postdoctoral fellowship. At the same time, we started to invite panelists from start-up biotech companies, Assistant professors from PUIs (Primarily Undergraduate Institutions), and journal editors. Diversifying the webinar topics and broadening the panelists helped us to regain the participants in our virtual webinars.
As a scientist, we perform experiments every day and learn through our mistakes and experiences. Organizing virtual events is a similar experience for Plant Postdocs as an organization. Over the two and half years, Plant Postdocs not only provided resources for the community, but also created a model system to follow and retain participants in this evolving era of virtual scientific events.
We believe the story of Plant Postdocs is an ideal example for non-profit scientific organization in this evolving era of science communication. We are committed to serve our community in future by providing resources, such as workshops, webinars, and training for ECRs, creating a platform for postdocs to present their work in our monthly seminar series, promoting pre-print and PREreview, and making the platform accessible to everyone.
Posted by Julian Lui, on 7 February 2022
During embryonic development, bone formation begins with the condensation of mesenchymal stem cells (MSCs). In a few places of our body, such as in the skull and the shoulder blades, mesenchymal condensations differentiate directly into bone-forming cells called osteoblasts. These osteoblasts make bones by directly laying down bone matrix, and this process is known as intramembranous ossification. In most other condensations in the embryo, such as those that eventually became the arms and legs, bones are formed by a different process called endochondral ossification, where a cartilage mold is first formed by chondrocytes, which is then replaced by the incoming osteoblasts, laying down bone matrix using the cartilage scaffold. Therefore, osteoblast function plays an important role in both types of ossification in our body and have clinical implications in both skeletal development and diseases like osteogenesis imperfecta and osteoporosis.
Runx2 and Osterix (also known as Sp7) are two of the most well-studied transcription factors that control osteoblast differentiation. Runx2 belongs to the Runx family that is composed of three genes, Runx1/Cbfa2, Runx2/Cbfa1, and Runx3/Cbfa3. Runx2 and 3 together are essential for chondrocyte maturation (1) and Runx2 is important also for osteoblast differentiation, as demonstrated by the complete lack of ossification in Runx2 knockout mice (2, 3). The molecular mechanisms by which Runx2 regulate osteoblast differentiation has been elucidated. All Runx2 family proteins contain a DNA-binding runt domain. The Runx proteins form heterodimers with transcriptional co-activator core binding factor b (Cbfb) in vitro (4) and specifically recognize a consensus sequence, PyGPyGGTPy (5), to upregulate a variety of osteoblast lineage-specific genes, such as Sp7 (osterix), Ocn (osteocalcin), and Bsp (bone sialoprotein) (6, 7). Similar to Runx2, Sp7 knockout mice also demonstrated lack of ossification. But in contrast to Runx2 knockout, the Sp7 knockout mice do express Runx2 in osteogenic cells, thus suggesting Sp7 acts downstream of Runx2 during osteoblast differentiation (8).
Over the years, the role of Sp7 in osteoblast function has been confirmed in humans. Pathogenic variants in SP7 have been described in patients with recessive osteogenesis imperfecta (OI type XII), which is characterized by generalized osteoporosis (9), and genome-wide association studies (GWAS) have identified common genetic variants in SP7 to be associated with bone mineral density in the general population (10). However, despite the physiological and clinical importance of SP7, much is yet to be learnt about its molecular mechanisms of action. Based on its similarities with other SP family members, SP7 was initially thought to bind GC-rich sequences (11). However, other studies have shown evidence for a lack of such binding preference for GC-rich sequences (12). In 2016, a study from Andrew McMahon’s group showed that SP7 differs from other SP proteins in that it has a lower binding affinity for GC-rich sequences, and instead preferentially binds to AT-rich sequences in osteoblast target genes through interactions with DLX proteins (13). These findings provided important insights into why SP7, but not other SP family proteins with similar zinc finger domains, is uniquely important for osteoblast function. However, these findings were based on in vitro chromatin immunoprecipitation sequencing (ChIP-Seq) using osteoblasts, and thus it remains unclear whether this unique AT-motif-binding of SP7 is important in vivo, as at least until a human subject is found to have such binding preference disrupted.
And that brings us to our work recently published in Nature Communications (14). In our latest study, we presented an Austrian boy with a complex skeletal disease which included craniosynostosis, severe scoliosis, long bone fragility, with areas of increased bone, particularly with thickened intramembranous bones, and other areas of decreased bone. In that patient, we identified a pathogenic heterozygous missense variant in SP7 (S309W). The complex skeletal phenotype and the apparently dominant nature of the variant differ markedly from the prior cases of SP7-associated recessive osteogenesis imperfecta and is not readily explained by a simple loss of SP7 function and osteoblast formation.
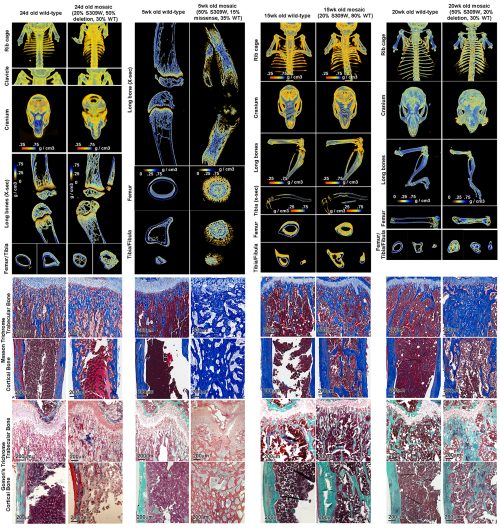
We created a mouse with the orthologous missense variant in Sp7, which partially recapitulated the human skeletal phenotype. Importantly, we showed evidence that the variant altered the binding specificity of SP7, with increased binding to GC-consensus sequences and decreased binding DLX proteins and to AT-rich motif (thus a neomorphic/gain-of-new-function mutation rather than a simple loss- or gain-of-function mutation). The variant tends to reverse the unique sequence specificity of SP7, causing it to revert to a specificity more similar to the other SP family members. Our current study therefore provides the first in vivo evidence that the unique AT-binding specificity of SP7 is indeed essential for normal bone development in vivo. Importantly, our study also suggests the possibility that other unresolved rare genetic disorders could also be caused by neomorphic mutations in transcriptional regulators.
Thank you for reading!
Posted by Aitana Castro, on 3 February 2022
The Quintay Practical Course in Developmental Biology 2020 student cohort interviewed Dr. Angela Nieto Toledano
Dr. Angela Nieto is a researcher, professor, scientific communicator whose work builds bridges between embryology and health research. Angela is the president of the ISDB (International Society of Developmental Biology), and she has received several scientific awards in recognition to her outstanding work – including her recent incorporation to the Royal Academy of Exact, Physical and Natural Sciences of Spain.
During the last MBL course in Quintay (January 2020), 18 students from the field of developmental biology had the opportunity to share meals, lectures, talks, ideas and ask advice from Dr. Nieto. Some of the discussions are summarized in this interview to share the experience more broadly with the developmental biology community.

1. What is snail and how did you start studying this gene? Why is snail so important for development and at the same time for processes involved in the evolution of cancer?
During my postdoc in London I had been working with David Wilkinson on the search for genes important for the development of the brain. Just before returning to Spain at the end of 1992, I found a new gene that was expressed in cells that were moving from one place to another in the embryo, a well-known process for which there was not much information on how it happened. We called that gene Slug, now known as Snail2, as we had previously isolated another family member, Snail1. Just then I decided that, in my new lab in Spain, I would devote my efforts to finding out how cells in the embryo move to form organs. In 1994, we found that Snail2 downregulation in chicken embryos prevented migration of both the neural crest and the mesoderm from the neural tube and the primitive streak, respectively. In this paper, published in Science, we hypothesized that the pathological activation of these genes could be involved in the acquisition of migratory and invasive properties in cancer cells, as at the cellular level, the delamination of cells from the primary tumour to start the metastatic cascade looked very similar to the delamination of neural crest cells from the neural tube. In 2000, we could show that this was the case.
2. What other things has your lab been interested in over the years?
We have continued to study the mechanisms that drive morphogenesis. Over the years we have found that the mechanisms that should only be active in the embryo are activated again in an aberrant way in different diseases, including cancer or organ degeneration. We continue to learn from the embryos in order to better understand the diseases and propose better therapeutic strategies.
3. Do you think that, in addition to effort and good ideas, there are rules to achieve success in a project? What are yours?
Passion, enthusiasm and resilience. It is also important to mention that scientific activity has degrees of freedom that other jobs do not have and this is very satisfying.
4. Your laboratory, like many in today’s world, is made up of researchers and students from multiple backgrounds. What are the advantages of this diversity?
In the lab, like in other aspects of our life and in nature in general, diversity is a source of richness and a selective advantage. A multicultural environment fosters knowledge and generosity and, in any case, science is universal and should promote strong links among individuals. We are now in the middle of a devastating and unexpected crisis, and we know that we will overcome Covid-19 only working all together. Scientific research has now taken centre stage in the news, let´s hope that it will continue to be, firstly because scientists could find a treatment and an efficient vaccine, and secondly because society realized the importance of investing in science as the only way to secure a prosperous future.
5. What advice would you give to female young researchers who want to develop a prolific career?
The career of a scientist is not easy, but in the case of women it is even more complicated because the crucial stage when the future is decided coincides with that of motherhood. We are still a long way from women being able to reconcile both aspects naturally. On the other hand, there are fields of science that have been and still are associated with men, and this means that many girls do not have female role models to look at. Girls develop the so called “dream gap”, something like the conviction that they are not going to be able to do well in scientific disciplines. This is now more dramatic than ever, as we are in a world of Technology, Artificial Intelligence and “Big Data”, where it is predicted that 80% of future jobs will be in the STEM disciplines. Therefore, we must help in two ways. The first is to help women who want to be scientists to continue their careers while they are mothers and the second is to change the sexist education that girls receive from the beginning. To do this, we must show models of successful women so that they see that it is possible and that they can do it too. In my case, I have been very lucky because I have always had the support of my family and my partner, and I have not had episodes of discrimination as I have progressed in my career. But we still have a long way to go.
6. Finally, this international course is composed mostly of students from Latin America, where many people dream of working in science since elementary school but have not had the best opportunities. What is your message to them?
First of all, I have to say that I have been very much impressed by the knowledge and enthusiasm of the students, and it has been a real pleasure to spend these days in Quintay. Thus, I want to thank Roberto Mayor and the other organizers for their efforts in organizing such a wonderful course for the last 20 years (and the many to come). My message to the students is quite simple, think high and work hard. And also think that science is fun and that, little by little, maybe we can help to better understand disease and improve our future.
The next EMBO Practical Course on Developmental Biology in Quintay, Chile from 4-16 January 2023 is now accepting applications. You can find more information on the course and how to submit your application here.
Posted by Gabrielle Kardon, on 2 February 2022
Closing Date: 11 February 2022
Looking to conduct research in molecular biology and genetics? We are looking for two lab technicians/specialists to assist in research on muscle stem cells, development, regeneration, disease, and effects of viral infection. More details about our research can be found at http:// www.kardonlab.org/ Technician will assist in management of a mouse colony as well as conduct supervised research (leading to publications). Technician must be reliable, well organized, detail-oriented, excited about research and committed to working in our lab for at least two years. Prior lab experience is preferred (although not necessarily required), and class work in biology and enthusiasm for science is essential. Lab is located at the University of Utah in Salt Lake City, affording amazing opportunities for science and outdoor recreation. Excellent team environment with great benefits.
Looking for two technicians/specialists to start Feb-June 2022. Please contact Gabrielle Kardon (gkardon@genetics.utah.edu) with CV, list of references, and a brief statement about why you are interested in the position. BS or BA required. For full consideration, apply by Feb 11, 2022.
Posted by Dhruv Raina, on 31 January 2022
(or Navigating post-PhD turbulence in the midst of the pandemic)
Working in academic research was such a blast that I’d never imagined anything else for myself. After my Master’s degree in regenerative medicine, I worked at the National Center for Biological Sciences (NCBS) in Bangalore, India, for a few years before I came across a post on The Node advertising a terrific project in Dr. Christian Schröter’s lab at the Max Planck Institute of Molecular Physiology (MPI). I had a fantastic time working on my doctoral projects at Chris’ lab, and the years packed with long debates about robustness and heterogeneity, coupled with many wonderful lab outings (pre-pandemic, of course :’-[ ) simply flew by. Last year, after I handed in my thesis, I saw an ad for Mosa Meat, a start-up working on developing cultured meat and made what appeared to be a sudden decision to change my career trajectory; I decided to join them.

I’ve always been captivated by a common refrain in biology: the tension between the fidelity of information transmission required in a process and the stochastics-driven heterogeneity or noise at that operational scale. As my PhD on signalling and fate choices in mouse embryonic stem cells drew to a close, I had my eye out for other systems where I could continue to study different aspects of this tension between robustness and exploratory behaviour. At the same time, after the tumultuous events of recent years, the call-to-action for addressing the climate crisis and re-thinking modern supply chains had firmly lodged itself in my heart. I was craving an outlet for these energies.
My first exposure to cultured meat came from an unlikely source: my partner’s father. He mentioned cellular agriculture in a conversation we had about changing the way we eat and for weeks after, the idea kept playing over and over again in my mind. When I saw Mosa Meat was hiring, I reached out to Joshua Flack, the scientist who’d placed the ad, in a cautious email peppered with tentative phrasing, to enquire a bit more about the role. Among the puzzles they were trying to solve, one stood out to me clearly – the question of how to robustly guide cells into certain fates and overcome the hydra of heterogeneity. After a set of interviews and a few chats with Josh and the other brilliant scientists at Mosa Meat, I was sold – here was a problem that promised to be both intellectually challenging and morally fulfilling.

Mosa Meat, as a food technology start-up, is packed to the gills with engineers and biologists at varying levels of experience, all of whom are simultaneously working at cracking different parts of this complex puzzle: how do we create cleaner and kinder meat? Working as a senior scientist in the Stemness and Isolation team, I’m trying to figure out the cell biology of bovine satellite and fibro-adipogenic progenitor cells in our bioreactors, how to guide them to retain their stemness and, later, to differentiate. What does this mean in practice? My daily life here looks a lot like it looked back in academia but with some differences. The similarities between the two are all the necessary verbiage of science: cell culture, banging your head against the wall trying to design an experiment that feels neat, imaging, flow cytometry, grabbing someone in the hall to have a quick chat about their recent results, and so on. The only practical difference is that instead of the small teams that academic environments foster, everyone here works as one big team that is fully invested in each others’ work. This means that in addition to discussions over Slack and email, plenty of in-person meetings are held to make sure everyone is up-to-date on the most recent findings. It’s a bit of an unusual experience for me (and by no means one that’s unwanted!) to work in a team of over a hundred scientists and engineers who are all excited about your most recent experiment.
Satellite cells finding their way into a myofiber at Mosa Meat. Imaged on a Phasefocus Livecyte microscope
At a higher level of abstraction, there are more differences between my work at the MPI and the work I’m doing here at Mosa Meat. For me personally, the work I did at the MPI was very focused on pushing advances in conceptual frameworks, whereas the focus here is more on leveraging conceptual advances to develop robustness in method. I certainly don’t mean to paint these differences as a dichotomy, and while in any project most of us think along both these tracks simultaneously, at Mosa Meat there is a bit more emphasis on the latter.
When I now reflect on my professional trajectory over the past year, it doesn’t feel like a sudden change of career but more like a natural extension of my personality and interests. In the end, I found that once I had distilled my initial desire to work in academia down to a few core drivers, I no longer thought about my career choices framed as a debate between ‘industry vs. academia’. I realized that what I really wanted to do was work at the bleeding edge of discovery and to strike out in a new and relatively unexplored direction.
Packaged into this career choice was also an inevitable geographic relocation. I won’t say too much about how this factored into my decision, since it’s a subject everyone in science has to contend with, and how one deals with it is specific to the individuals and their circumstances. While moving countries in the middle of the pandemic and wrestling with all the ensuing paperwork was personally a hard decision (especially for us as immigrants), I was fortunate in that my partner (also a scientist) and I were deeply familiar with the infamous two-body problem as it relates to relationships and lab locations, and were well prepared for it.
After five months of working here in beautiful Maastricht with a wonderful team of very smart people who share similar ethics and values around sustainability as me, I still can’t wait to burst into the lab every day. It’s tremendously exciting to work at a start-up that is trying to simultaneously solve not just a difficult biological problem but also a tough engineering and scale-up challenge.
And yes, of course, I’m also driven by the temptation of getting my teeth into a delicious Mosa burger!
Posted by Douglas Houston, on 31 January 2022
Doing great science depends on teamwork, whether this is within the lab or in collaboration with other labs. However, sometimes the resources that support our work can be overlooked. In our new series, we aim to shine a light on these unsung heroes of the science world. The fourth article in the series is by Doug Houston (Interim Director, DSHB) who describes the work of the Developmental Studies Hybridoma Bank.
A short history of monoclonal antibodies and the DSHB
In 1975, Köhler and Milstein1 published a method for selecting and immortalizing cells that secreted a single, monospecific antibody, or monoclonal antibody. This advance ushered in the era of ‘biologics’, or biological molecules with exquisite specificity akin to chemical molecules. Monoclonal antibodies are made by fusing an antibody-producing mature B cell (usually obtained from the spleen of an immunized mouse) and an immortalized myeloma cell, producing a ‘hybridoma”. As the number of labs making and using these hybridomas grew, it became apparent that community repositories would be needed to effectively preserve and share these reagents.
The DSHB was one of these early repositories, and is one of the few, if not the only one, still in existence and operating independently. The DSHB was formed in 1986 through a contract with the National Institute of Child Health and Human Development (NICHD) involving Thomas August at Johns Hopkins University and Michael Solursh at the University of Iowa. Dr. August ran the Johns Hopkins portion, which generated new hybridomas, mainly focusing on immune cell development, while the University of Iowa group directed by Prof. Solursh in the Department of Biology stored and distributed hybridomas and antibodies.
Prof. Solursh’s lab was focused on limb development and early hybridoma deposits reflected these interests. Early hybridoma deposits expressed antibodies recognizing important molecules in limb development including muscle and matrix proteins. One of the first antibodies deposited was MF 20 (anti-sarcomeric myosin heavy chain), which was contributed by Donald Fischman in 1986. MF 20 remains one of the most requested and robust monoclonal antibodies in the DSHB collection. The skeletal muscle and extracellular matrix hybridoma collections grew substantially in the initial years. After Prof. Solursh tragically died at a young age in 1994, the Iowa division of the DSHB came under the interim directorship of David Soll, and daily operations were run by Karen Jensen, a long-time research associate in the Solursh Lab. As the collection of hybridomas in Iowa grew and the depth of curation and expertise surrounding these antibodies became established, the DSHB was consolidated at Iowa under Prof. Soll’s directorship.
In 1998, the NICHD’s contract with the DSHB was ended (three years ahead of schedule) and the Bank has been independent of NIH and self-supported ever since. Our operating expenses are covered by distribution fees, which are kept as low as possible (at cost) to facilitate wide use of the antibodies/hybridomas by the worldwide scientific research community. The DSHB still maintains close relationships with various NIH entities, including the National Cancer Institute (NCI) and the Common Fund. The DSHB is the preferred distributor for NIH, HHMI and MDA funded monoclonal antibody efforts (among others).
The people behind the DSHB
The DSHB is run by a relatively small but dedicated group of office staff and scientists. Our staff are experts at navigating the changing rules of university policies, shipping and customs rules, and addressing the concerns of researchers from around the globe. Many have been with the Bank for over a decade and a select few have worked with the DSHB since its inception. Notably, Dr. Karla Daniels (the voice on the phone when you call us!) worked with Profs. Solursh and Soll and has defined the role of our senior scientific curator of the DSHB collection and expert technical advisor, disseminating information on best use of our antibodies. Brian and Rebecca maintain the hybridomas cell lines and produce the antibodies we distribute. Working with hybridomas is an artisanal science, and Brian and Rebecca have mastered the art of coaxing these cells out of cryostorage and getting them to produce large quantities of immunoglobins. Nicki, Mitch and Brian process and manage the orders and track them until delivered to the destination labs. We’ve recently hired a dynamic new cohort of staff including Nick, Mejd and Nisha to continue the tradition of high-quality antibody production and service
What is available for researchers?
The DSHB provides two valuable services for researchers. First, scientists can ‘deposit’ their hybridomas (or antibodies) with the DSHB, granting the DSHB non-exclusive rights to distribute the antibody products as well the hybridoma cell lines (if desired) to the scientific research community. The depositor and the originating institution retain the intellectual property to the hybridoma and thus can benefit from commercialization while still making the reagent available to researchers. Second, researchers can purchase antibodies contributed by other scientists, with the stipulations that the reagents be used for non-commercial research or teaching and not be transferred or reverse engineered. The original mandate of the DSHB was to keep the costs to researchers as low as possible, and we continually strive to be the main provider of high-quality but affordable antibodies. Researchers typically purchase hybridoma-conditioned cell culture supernatant to use in immunostaining or immunoblotting, although other higher concentration options such as bioreactor supernatants or concentrated supernatants are available. Custom orders for large amounts of antibody or purified immunoglobin may also be requested.
We also accept deposits of polyclonal antisera, and although it has not been widely advertised (yet!), the DSHB is also accepting plasmids encoding recombinant antibodies and distributing the antibodies produced by transfected cells (DSHB will not distribute the plasmids themselves). These recombinant antibodies are an important recent advance in antibody technology and are the main way to produce monoclonal antibodies made in rabbits. Rabbit antibodies (traditionally used as polyclonal antisera) have different properties from those of mice, including better responses to shorter epitopes and a more diverse immune response2. Rabbits may thus be better for making antibodies against phospho- or other modified amino acid epitopes and represent an important complement to traditional mouse monoclonals.
How can the community contribute?
Because the DSHB considers itself a community resource, the research community provides critical feedback and support for the DSHB. Scientists can help support the DSHB by contributing hybridomas and/or antibodies when appropriate. It’s humbling to see the list of prominent scientists who have contributed reagents (see the ‘contributor spotlight’ on the DSHB website: https://dshb.biology.uiowa.edu). Also, the DSHB and other repositories can be supported by purchasing from them whenever possible. The DSHB and other repositories are critical for fostering rigor and reproducibility in research, a goal promoted by NIH and other funding agencies. Depositing with the DSHB is thus an easy way to implement ‘resource sharing plans’ in grant applications.
Because part of our mission is to keep costs low, the DSHB (unfortunately) does not actively characterize or validate each antibody, but depend on the depositor’s validation. Validation builds as each antibody accrues citations and confirming applications. Users can contribute by sending feedback about antibodies received from the DSHB. The low cost of DSHB is advantageous for experimenting with different conditions and antibodies, and we hope that labs tell us what works and what doesn’t! As community efforts directed at validating monoclonal antibodies improve (do they specifically recognize the target antigen, and only the target antigen?), we hope to improve access to this information for our collection. Users can always send us images of the antibodies at work, and if we can’t use them on the web site, we’ll disseminate on social media (Twitter: @_DSHB_; FB: https://www.facebook.com/DSHBUI/).
And last, as always, please cite the antibodies and contributors of useful antibodies. This is especially important for new or untried antibodies; many of these are potentially useful but may not be widely adopted unless cited in the literature.
User comments
Many antibodies at the DSHB are unique, absent from the catalogues of large antibody companies, and we often hear from customers how antibodies in the DSHB collection are essential to their research. This is especially true for those working in model organisms where commonly available antibodies made against human and mouse homologues may not work. Many commenters wish we had more antibodies in our collection. We also receive comments appreciative of our customer service. Of course, we receive the ‘rare’ complaint too. We actively troubleshoot use of the antibodies, drawing on the expertise of our scientists, who have not only deep general knowledge, but often long histories of working specifically with the antibodies in question over the years. Our favourite comment however is from someone who wished we were a hundred times bigger!
We hold this as an aspirational goal. If money were no object, we would exponentially grow the number of antibodies to human proteins and establish a robust validation pipeline. But regardless, the DSHB will continue to foster the development and sharing of antibodies for model (and non-model) organisms, which have contributed the most to the discovery of new biological insights.
Any hidden gems, features that are new, or that researchers might be less aware of?
The DSHB collection has continued to grow, from several hundred in the 1980s -1990s to over 5000 currently. Recent additions include a large number of recombinant antibodies from the Clinical Proteomic Technologies for Cancer program of the National Cancer Institute, which includes over 800 antibodies against cancer-related targets. These include many antibodies related to DNA damage repair, RAS/MAPK signalling, including regulatory phospho-epitopes, as well many transcription factors involved in epithelial-mesenchymal transition. Through the Protein Capture Reagent program of the NIH Director’s Common Fund, we took deposit of over 700 transcription factor antibodies, optimized for ChIP3. These were recently evaluated for ChIP-exo/seq by Frank Pugh’s group at Penn State University, in a recent publication, and many antibodies gave superior results using just ‘raw’ antibody-containing supernatant4. Also, last year, the NeuroMab collection of neuroscience and neurodevelopment antibodies (over 500 hybridomas), developed by James Trimmer at UC Davis5, was deposited with the DSHB. There are doubtless many hidden gems among the uncharacterized antibodies at the DSHB, although we think the DSHB itself is also a hidden gem, and we are grateful to the Node for featuring us and other science resources!
Douglas W. Houston
Interim Director, DSHB
Iowa City, IA. Jan. 2022
1Köhler, G. and Milstein, C. (1975). Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497.
2Weber, J., Peng, H. and Rader, C. (2017). From rabbit antibody repertoires to rabbit monoclonal antibodies. Exp Mol Medicine 49, e305.
3Venkataraman, A., Yang, K., Irizarry, J., Mackiewicz, M., Mita, P., Kuang, Z., Xue, L., Ghosh, D., Liu, S., Ramos, P., et al. (2018). A toolbox of immunoprecipitation-grade monoclonal antibodies to human transcription factors. Nature methods 15, 330–338.
4Lai, W. K. M., Mariani, L., Rothschild, G., Smith, E. R., Venters, B. J., Blanda, T. R., Kuntala, P. K., Bocklund, K., Mairose, J., Dweikat, S. N., et al. (2021). A ChIP-exo screen of 887 PCRP transcription factor antibodies in human cells. Genome Research 31:1663-1679.
5Gong, B., Murray, K. D. and Trimmer, J. S. (2016). Developing high-quality mouse monoclonal antibodies for neuroscience research – approaches, perspectives and opportunities. New Biotechnol 33, 551–564