Looking to crustaceans to understand insect wing evolution
Posted by Courtney Clark-Hachtel, on 19 October 2020
The wings of vertebrates, like birds and bats, emerged relatively recently, and we understand that these wings evolved from forelimbs. Even for the mythological dragon there seems to be a consensus (at least in recent depictions) that their wings are also derived from their forelimbs. Insects, however, possess both wings AND limbs on their winged segments, suggesting that their wings are not evolved from modified limbs, which begs the question “where did insect wings come from?” Despite their status as the first animals to take to the skies, the answer to this question has remained poorly understood and under debate for over 200 years1,2. This debate has culminated in two leading hypotheses on the origin of insect wings, each linked to different origin tissues: a lateral outgrowth of the dorsal body wall (tergum) and ancestral proximal leg structures (pleuron in insects)2.
Historically, scientists have attempted to dissect the evolutionary origin of insect wings through identifying structures related to wings in non-winged segments of insects (wing serial homologs) and other wingless arthropods (wing homologs)3. The idea behind this approach is that the wing homologs in other segments have been modified to differing degrees, suggesting that wing homologs from wingless segments might provide us with a series of “snapshot” views into the evolution of wings and help us reconstruct how this complex structure came to be. Until relatively recently, attempts to identify wing-related structures in non-winged segments relied on these structures sharing morphological similarity with wings (i.e. looking like wings), which understandably limited the identification of wing homologs. Recently, with the application of molecular evolutionary and developmental biology (evo-devo) approaches, the diversity of the tissues identified as wing homologs has increased3. Some of these studies have even provided evidence that insect wings are not derived from either tergum or pleuron (ancestral proximal leg), but potentially from a combination of these two tissues (i.e. they have a dual origin)4–9.
In our recent paper, we applied this molecular evo-devo approach to the identification of wing-related structures in a crustacean10. The reasoning behind looking for “wings” in a crustacean is that crustaceans and insects share a common ancestor. Therefore, by identifying the potential wing homologs of a crustacean and comparing them to the wing serial homologs of insects, we can gain a better understanding of what tissues were present in the common ancestor of these groups that had the potential to become wings in insects. Parhyale hawaiensis, the crustacean we chose for our study (Fig 1), provided a great “model” crustacean for our investigation because their dorso-ventral body plan is very similar to that of insects, which makes comparisons between the two much simpler.

We started our search for wing homologs in Parhyale by identifying structures that are dependent on the gene vestigial (vg). vg is a critical wing gene in insects and has often been used for the molecular identification of wing homologs. We looked at expression and function of vg in Parhyale and noticed something striking. First, vg is expressed in both the tergal edge and the proximal leg segments. Second, when we knocked-out the function of vg via CRISPR/Cas9 genome editing, we saw that the development of BOTH of these tissues (tergum and proximal leg, related to insect pleuron11) was disrupted (Fig 2). We expanded our expression and function analyses to two more insect wing genes, nubbin (nub) and apterous (ap) and saw a similar outcome – these genes are expressed and/or function in both tergum and proximal leg segments (Fig 2). We were curious how big the overlap was between genes that function in wing development (wing gene network, WGN) and genes that function in tergum and proximal leg development of crustaceans, so we investigated the expression of a few additional wing genes in Parhyale. Many of these genes were also expressed in the tergum and proximal leg of Parhyale, and one of these genes, optomotor-blind (omb), showed impressive expression pattern overlap with the functional and expression domains of vg, nub, and ap (Fig 2).
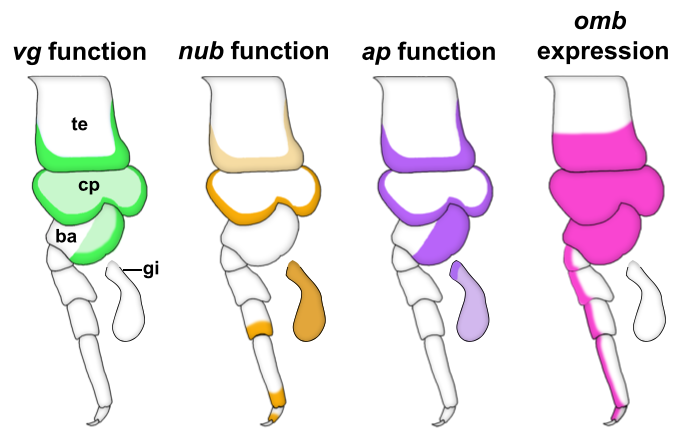
Through these investigations, we were able to show that a gene network similar to the WGN operates in both the tergal edge and proximal leg of Parhyale, suggesting that the evolution of this network precedes the emergence of insect wings. It also seems that both of these structures qualify as candidates for wing homologs of a crustacean. When we compare these structures to those that have been identified as wing serial homologs in the wingless segments of insects, we see a striking similarity; two separate tissues dependent on wing genes, one of tergal and one of pleural/proximal leg-related identity (Fig 3). The similarity between the wing-related structures in insects and crustaceans appears to point to a dual evolutionary origin of the insect wing and suggests that insect wings evolved through the merger of two previously distinct structures.
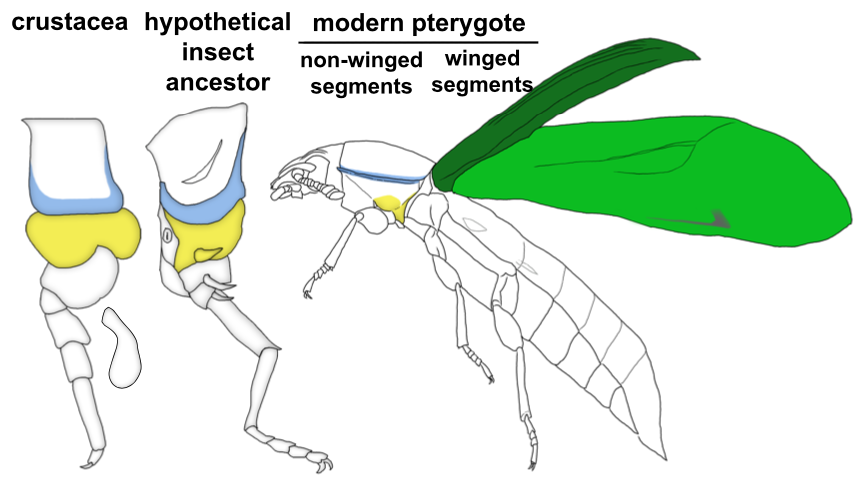
An aside about terminology: Historically, tissues related to wings on non-winged segments (either in insects or crustaceans) have been referred to as “wing homologs”. We are starting to see that this terminology is problematic especially when you consider the evolutionary order of the emergence of these structures. It seems that the “ancestral state” for wings is really two separate, previously existing structures in the form of tergum and pleuron (or ancestral proximal leg segments). Only in the winged segments of insects do these two previously separate structures seemingly merge to form the wing (Fig 3). Therefore, more accurate terminology for the wing-related structures on non-winged segments might instead be “tergal serial homologs” or “pleural serial homologs” as this is more representative of the ancestral state for these tissues. After all, it is becoming increasingly apparent with recent studies that these “tergal” and “pleural” serial homologs provide an “evolutionary hotspot” for the development of morphological novelties including, but not limited to, beetle thoracic horns, tree hopper helmets, beetle abdominal gin traps, and wings12.
References:
- Grimaldi, D. & Engels, M. S. Insects take to the skies. in Evolution of the Insects 155–187 (Cambridge University Press, 2005).
- Clark-Hachtel, C. M. & Tomoyasu, Y. Exploring the origin of insect wings from an evo-devo perspective. Curr. Opin. Insect Sci. 13, 77–85 (2016).
- Tomoyasu, Y., Ohde, T. & Clark-Hachtel, C. M. What serial homologs can tell us about the origin of insect wings. F1000Research 6, 268 (2017).
- Clark-Hachtel, C. M., Linz, D. M. & Tomoyasu, Y. Insights into insect wing origin provided by functional analysis of vestigial in the red flour beetle, Tribolium castaneum. Proc. Natl. Acad. Sci. U. S. A. 110, 16951–16956 (2013).
- Medved, V. et al. Origin and diversification of wings: Insights from a neopteran insect. Proc. Natl. Acad. Sci. U. S. A. 112, 15946–15951 (2015).
- Elias-Neto, M. & Belles, X. Tergal and pleural structures contribute to the formation of ectopic prothoracic wings in cockroaches. R. Soc. Open Sci. 3, 160347 (2016).
- Linz, D. M. & Tomoyasu, Y. Dual evolutionary origin of insect wings supported by an investigation of the abdominal wing serial homologs in Tribolium. Proc. Natl. Acad. Sci. U. S. A. 115, E658–E667 (2018).
- Tomoyasu, Y. Evo–devo: The double identity of Iisect wings. Curr. Biol. 28, R75–R77 (2018).
- Clark-Hachtel, C. M., Moe, M. R. & Tomoyasu, Y. Detailed analysis of the prothoracic tissues transforming into wings in the Cephalothorax mutants of the Tribolium beetle. Arthropod Struct. Dev. 47, 352–361 (2018).
- Clark-Hachtel, C. M. & Tomoyasu, Y. Two sets of candidate crustacean wing homologues and their implication for the origin of insect wings. Nat. Ecol. Evol. (2020). doi:10.1038/s41559-020-1257-8
- Bruce, H. S. & Patel, N. H. Insect wings and body wall evolved from ancient leg segments. bioRxiv (2018). doi:10.1101/244541
- Linz, D. M., Hu, Y. & Moczek, A. P. From descent with modification to the origins of novelty. Zoology 143, 125836 (2020).


 (4 votes)
(4 votes)
 (No Ratings Yet)
(No Ratings Yet) (9 votes)
(9 votes)


 Features of our program include:
Features of our program include: