Future developments: your thoughts and our plans
Posted by Katherine Brown, on 5 January 2016
This editorial by Olivier Pourquié and Katherine Brown first appeared in Development.
As a journal with its community very much at its heart, we here at Development believe it is essential to ensure we take your opinions into account when planning for the future. It was with this ethos in mind that we recently carried out a community survey looking at how well the journal reflects the current state and future directions of the field. Firstly, we’d like to take this opportunity to thank all of you who took the time to complete the survey – we were overwhelmed by the number of responses and by the detailed feedback we received. We’d like to report back on the results of that survey and on how these are now influencing the journal’s plans.
Developmental biology as a field is evolving. While these aren’t necessarily easy times for fundamental research, with funding tight and an ever-increasing pressure for work to have ‘translational impact’, we would argue that they are exciting times for our field. Genome sequencing and editing technologies have opened up a world of non-traditional model systems to genetic analysis, allowing us to probe the evolutionary and developmental mechanisms underlying the development of a wide range of species. Quantitative and computational techniques allow us to probe these mechanisms in greater detail. In vitro technologies using stem cells provide tools to analyse fundamental principles and mechanisms of cell proliferation, differentiation and even morphogenesis. These techniques are proving to be particularly important for providing insights into human development, which can not be addressed in vivo. Development’s mission is to publish the best research across the scope of our field: to publish papers at the cutting edge of modern developmental biology, as well as continuing to be the natural home for the best work in more traditional areas.
So, how well are we succeeding in that goal, and how well do you – our community – think we are doing? These were some of the questions we set out to explore with our recent survey, which gathered over 700 responses and included lots of detailed comments (see Box 1 for some of the key statistics). We’re still sifting through some of the data, but this has given us a very valuable impression of where you think Development sits within the publishing landscape. In general, you told us that the journal reflects our field very well. While a few of you raised some concern about a perceived decline in quality or influence, others told us that Development is the premier journal for the field and a key source of essential reading matter. We asked whether particular fields were over- or under-represented in the journal. Most of you think we are doing a good job in covering a broad spectrum of areas, although your comments have highlighted particular areas (such as quantitative, computational and systems biology, ‘omics, evo-devo and plant development) where perhaps we need to do more to attract more high-quality submissions. We are looking at ways of doing this and are, for example, in the process of putting together a Special Issue on Plant Development, which is still open for submissions (see http://dev.biologists.org/content/special-issue-plant-development). Some of you feel that we are publishing too much stem cell work (more on which below) or that too many of our papers use too narrow a range of model systems. But in general, it seems that although there are always improvements to be made, you feel that the papers we publish reflect the diversity of research being carried out in our community.
————————————————————————————————————————————————————
Box 1. Key survey responses
Question: Considering the content of Development over the past year or so, how well do you feel the journal reflects the state of the modern developmental biology field?
Rate on a scale of 1 (not at all well) to 5 (very well)
Average score: 4.15
Question: Please rate the extent to which you think changing Development’s name (e.g. to Development and Stem Cells or similar) could benefit/disadvantage the journal.
Strong/slight disadvantage: 65%; Neutral: 21%; Strong/slight advantage: 14%
Question: Please rate the extent to which you agree/disagree with the following statements (only selected statements shown):
‘I am supportive of Development’s move into the stem cell field and I think the balance of content is about right.’
Agree: 48%; Neutral: 27%; Disagree 25%
‘I believe there is space for some stem cell papers in the journal, but only where there is a direct link to an in vivo developmental process.’
Agree: 72%; Neutral: 15%; Disagree 13%
‘I feel that stem cell papers do not belong in a developmental biology journal.’
Agree: 11%; Neutral: 17%; Disagree 72%
————————————————————————————————————————————————————
The second part of the survey focussed on our recent expansion into the stem cell field. This has been one of our major strategies over the past few years, and one that we have discussed extensively in previous editorials (e.g. Pourquié et al., 2013). We continue to believe strongly that this is an important direction for the journal (and for the field more generally), but we wanted to gauge opinions in the community and to ensure that we have your support in this. As we expected, responses here were quite polarised. The vast majority of you agree that Development should be publishing stem cell papers – particularly those where there is a clear link to an in vivo developmental biology process – and most of you feel that we have the balance of content about right. Your comments indicate that you, like us, recognise the deep connections between the two fields, and we are heartened to see that we have your support. That said, some of you expressed strong opposition to this move, arguing that most stem cell research isn’t really developmental biology, that stem cells are a ‘hot topic’ right now but that this is a passing fad, or that there are plenty of other stem cell journals out there and that Development should stick to core aspects of developmental biology. While we respect these opinions, our viewpoint is rather different. Clearly there are aspects of stem cell research, particularly the more translational work, that do not directly relate to developmental biology, and these do not belong in the pages of Development. Still, much of stem cell research, even the exclusively in vitro work, is clearly informed by developmental principles, or contributes to our understanding of these principles. The stem cell field is here to stay, although it will – like developmental biology – evolve over time, and we see an important role for Development in bridging the gap between these two interlinked fields.
We will therefore continue to strengthen our efforts to attract the best stem cell research to the journal. In particular, we are very keen to publish more research on human development (a field that several of you told us was under-represented in our pages). Our 2015 Special Issue on Human Development (see Pourquié, 2015) showcased some fantastic research in this field, and we have recently attracted more submissions in this area – a trend we hope we will continue. To this end, we are organising a meeting on this topic (a follow-up to our highly successful 2014 event) for September 2016; for more details, see http://www.biologists.com/meetings/from-stem-cells-to-human-development-2016/. We are also delighted to announce that we have recruited two new editors to Development’s editorial team. Paola Arlotta has joined the team to replace Magdalena Götz, who stepped down last year and whom we thank for her dedicated service to the journal. Paola, who is based at Harvard University (USA), is an expert in cortical neurogenesis, with a strong interest in how neuronal progenitor fate is regulated and how our understanding of these processes might impact on potential regenerative therapies for nervous system disorders. Rong Li steps down from the editorial team at the end of 2015, and we are also very grateful for her contribution over the past five years. Rather than recruit a direct replacement for Rong, we have instead decided to create a new ‘Guest Editor’ position. This temporary position will allow us to focus on a particular field of interest, and will be used over time to promote different up-and-coming areas of developmental biology. In keeping with our strategy to promote the analysis of developmental mechanisms using stem cell systems, our first Guest Editor will be Melissa Little (Murdoch Childrens Research Institute, Australia). Melissa has a long-standing interest in kidney development, and is now pioneering research into generating kidney tissues in vitro from human stem cells. We are absolutely delighted to welcome both Paola and Melissa to the editorial team.
Returning to the survey, perhaps the most controversial part asked whether Development should consider changing its name to reflect the inclusion of stem cell research. This is something we had been discussing internally – as it might help us to attract more and higher quality submissions from the stem cell community, as well as more accurately echo the current content of the journal – but without a clear consensus among the editorial team we wanted to gauge the opinion of the broader community. A clear majority of you said that changing our name would disadvantage the journal. While most of you responded that any potential name change would not affect the likelihood of your submitting to, reviewing for or reading Development, it is clear that our community wants Development to remain ‘Development’. And so it shall.
What other lessons have we learned from this survey? Interestingly, it seems that opinions about the journal are remarkably consistent across the age and career stage range. We already knew that many more established members of our community recognise the value of a journal like Development, but it was particularly pleasing to see that most early career scientists do too. We hope this stands us in good stead for the future, as those students and postdocs progress through their career. We also saw that, while many of you know about our publisher – The Company of Biologists – and its not-for-profit status and charitable activities, a sizeable proportion of the community does not know much about the organisation behind the journal. For those of you interested in finding out more, please see our recent editorial (Pourquié et al., 2015). Finally, and perhaps most importantly, we learned (from the size and nature of the response to this survey) just how much you care about Development. Your continued support and engagement is what keeps Development the important journal that it is, and we are hugely grateful to have it.
We would like to close by thanking all those who contribute to Development’s success. Our editors, editorial board members, staff and authors gain public recognition, but the importance of our dedicated referees should also be acknowledged. All those who reviewed for the journal in the past 12 months are listed in the supplementary information, and we are truly thankful for their time and dedication that ensures the quality of the work we publish.
References
- Pourquié, O. (2015). Human development: a Special Issue. Development 142, 3071–3072doi:10.1242/dev.129767.
- Pourquié, O.,Bruneau, B.,Götz, M.,Keller, G. and Smith, A. (2013). Stem cells and regeneration: a Special Issue. Development 140, 2445 doi:10.1242/dev.098350.
- Pourquié, O.,Brown, K. and Moulton, C. (2015). Developing a new look. Development 142, 3803–3804doi:10.1242/dev.131979.


 (2 votes)
(2 votes)



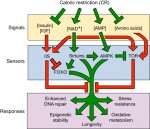 All multicellular organisms undergo a decline in tissue and organ function as they age. Here, Michael Schultz and David Sinclair discuss recent advances in our understanding of why adult stem cells age and how this aging impacts diseases, lifespan and potential therapies. See the Review on p.
All multicellular organisms undergo a decline in tissue and organ function as they age. Here, Michael Schultz and David Sinclair discuss recent advances in our understanding of why adult stem cells age and how this aging impacts diseases, lifespan and potential therapies. See the Review on p.  (No Ratings Yet)
(No Ratings Yet) 1-
1-