I have been a postdoc in London, alas at King’s College London (more on the reason for this regret in future), for 5 years now. There are some great things about London that overcome the horrendous prices and the relentless advance of painfully (un)cool hipster culture. If you are a bit of an obsessive developmental biologist, then YEM is one of them. Started by PhD students at UCL just before my time, this meeting, held with the kind support of Yoshiyuki Yamamoto (‘Yama’ to his friends and colleagues), I am reliably informed was initially nothing more than a geeks’ talking shop. The 2014 edition happened on the 27th June. It was massive. Delegates travelled from as far afield as Mill Hill. That is genuinely (I looked it up) in zone 4. There were even rumours that one of the speakers was from some obscure further education college near David Cameron’s home village.
Anyway, enough of the bad digression. The meeting has grown each year and now seems almost as large as a national society conference. This is to the enormous credit of the founders, the participants, but perhaps most of all, the organisers. To summarise, it was ace. As such, I thought I would post a very unofficial review. I have only picked talks that I particularly felt strongly about/enjoyed/didn’t fall asleep in, so my apologies to those I don’t mention. I have many anachronistic (many would say at best out-of-date and at worst downright ignorant) scientific views, so you shouldn’t feel at all slighted. Any opinions that follow are entirely my own and any disgruntled speakers, please find me at a conference where I will be happy to apologise and, if necessary, lubricate the apology with beer.
Dang Vinh Do (NUS & Cambridge)
We kick off (sorry, I was writing this during the World Cup though) with very early mammalian development, specifically the specification of lineages within the early mammalian blastocyst. This is a fascinating topic and one that has received huge attention over the last few years, not least because of the obviously huge potential medical implications. In the first talk, Dang Vinh Do described his work outlining the role of the JAK-STAT signalling pathway in maintaining the fate of the Inner Cell Mass that will form the embryo proper. This sounds a simple story, but it actually involved some really elegant genetics. The authors demonstrated that STAT3 was actually expressed maternally (in the egg). This nicely explained a paradox in the field: STAT3 was known to be crucial for embryonic stem cells in culture, but the knockout mouse made it to e6.5. What’s going on? To nail this contention that it was the maternal expression that was crucial, Dang Vinh Do ingeniously crossed female knockouts to male heterozygotes, and recapitulated the ESC phenotype. In his mice, the ICM was formed, but then subsequently Oct4 (one of the Yamanaka factors crucial for its development) disappeared and Cdx2 (a marker of the trophoectoderm) expanded. The mice made an embryo, but then lost it. Paradox solved. Dang has now (I think) moved to the germ cell lineage and to that technical college in East Anglia. I expect great things.
Mubeen Goolam (Cambridge)
Moving along in development, Mubeen Goolam (form a different lab in that technical college in East Anglia) spoke about his doctoral work examining Satb1, which is expressed preferentially in the ‘inner’ cells at the 16-cell stage of the developing embryo. Mirroring the situation with STAT3, a clear ES cell phenotype where Satb1 is required for differentiation into primitive endoderm is not revealed in the knockout mouse, which dies postnatally. Again, the answer was maternal expression, which when knocked down with siRNA yields a decreased amount of primitive endoderm, as judged by Sox17 expression. Now, a big theme in vertebrate development is the functional redundancy between paralagous developmental genes (a hangover from the whole genome duplications that kicked off vertebrate evolution in a lineage of unimpressive tiny chordates swimming around in the sea some 500 million years ago). However, remarkably in this case, the paralogue of Satb1, Satb2 has an antagonistic function – the satb1 knockdown phenotype is rescued by knocking down satb2. Not for the first time recently, I am left with the abiding feeling that early mouse development is profoundly unusual.
Erica Namigai (Oxford)
Staying on very early development (I do have other interests) Erica Namigai presented her doctoral work on the marine worm Pomatoceros lamarckii. This work, for me, highlighted the folly of concentrating scientific work on a small number of model organisms and the possibilities that accompany scientific decisions based solely on curiosity. There are basically no experimental techniques available in annelid worms. None. Well, you can chuck drugs on the embryos and see what happens, but for the moment that’s it. However, Erica’s work highlights the value of looking at things very carefully. In characterising early development, she noticed that remarkably (to my knowledge anyway) the second cell division is not coordinated. In 38% of embryos, there is a 3-cell stage. Now, this might just illustrate that Pomatoceros is not very good at developing under experimental conditions, but it set Erica thinking about how chirality is generated – how you make a left-right axis and break bilateral symmetry. Using some very trendy fluorescent labelling of vesicles as a way to visualise the cytoskeleton with light sheet microscopy, Erica showed that even at the two-cell stage, there is asymmetric distribution of cellular components that prefigures the eight cell stage when left-right asymmetry had previously been known to be established. Thus, in contrast to vertebrates and insects, the left-right axis can be set incredibly early and can in fact be the first body axis to form, not the last.
Daniele Soroidoni (Mill Hill)
Now, as I said, I am quite old-fashioned in some ways. I think, as I once heard Peter Lawrence say (at the first YEM I went to) that it’s really important to spend a lot of time just looking at stuff. Danielle Soroidoni, examining somitogenesis, did exactly this by taking advantage of the beautiful live imaging possible in the zebrafish. He generated a series of complex mathematical model describing the process and showing that the somitogenesis ‘clock’ was not in fact a clock and wave but a couple of waves with clockish oscillations that varied in amplitude across the anterior-posterior axis and eventually caught up with one another. I must confess, I finished feeling rather disorientated. In fact, my gathering thoughts were summed up by a questioner who posed a wonderful question, albeit a little bit confrontationally (I am paraphrasing slightly): ‘’does your descriptive analyses, however sophisticated and beautiful, tell us anything beyond the fact that the use of the word ‘clock’ in ‘segmentation clock’ is simply a metaphor?’’ Now, my sympathy is absolutely with the questioner (I like molecular mechanisms), but this kind of scientific difference of opinion (and even philosophy?) is absolutely why this kind of elegant descriptive work should exist. It makes people think.
Joseph Grice (Mill Hill)
The last talk I will mention is that by Joe Grice, which I loved. Despite not being able to turn on the computer (and being terribly English and affable about the whole thing – in my notes I wrote ‘I like him already!’), he nevertheless proceeding to give a fascinating talk using high-end comparative genomics. Through a combination of genomic analysis and transgenic assays in the zebrafish, he identified Hox/meis site combinations in regulatory elements that drive segment-specific expression in the vertebrate hindbrain. It is thus likely that these elements facilitated the evolution of such segmental gene expression patterns, linking the pre-existing Hox code with newly evolved segmentation in the hindbrain. Like the meeting as a whole, terrific stuff.
 (2 votes)
(2 votes)
 Loading...
Loading...
 Research and news:
Research and news: – Simon wrote about Microscopes4Schools, an outreach project that aims to bring microscopy to the classroom.
– Simon wrote about Microscopes4Schools, an outreach project that aims to bring microscopy to the classroom.

 (1 votes)
(1 votes) (2 votes)
(2 votes)

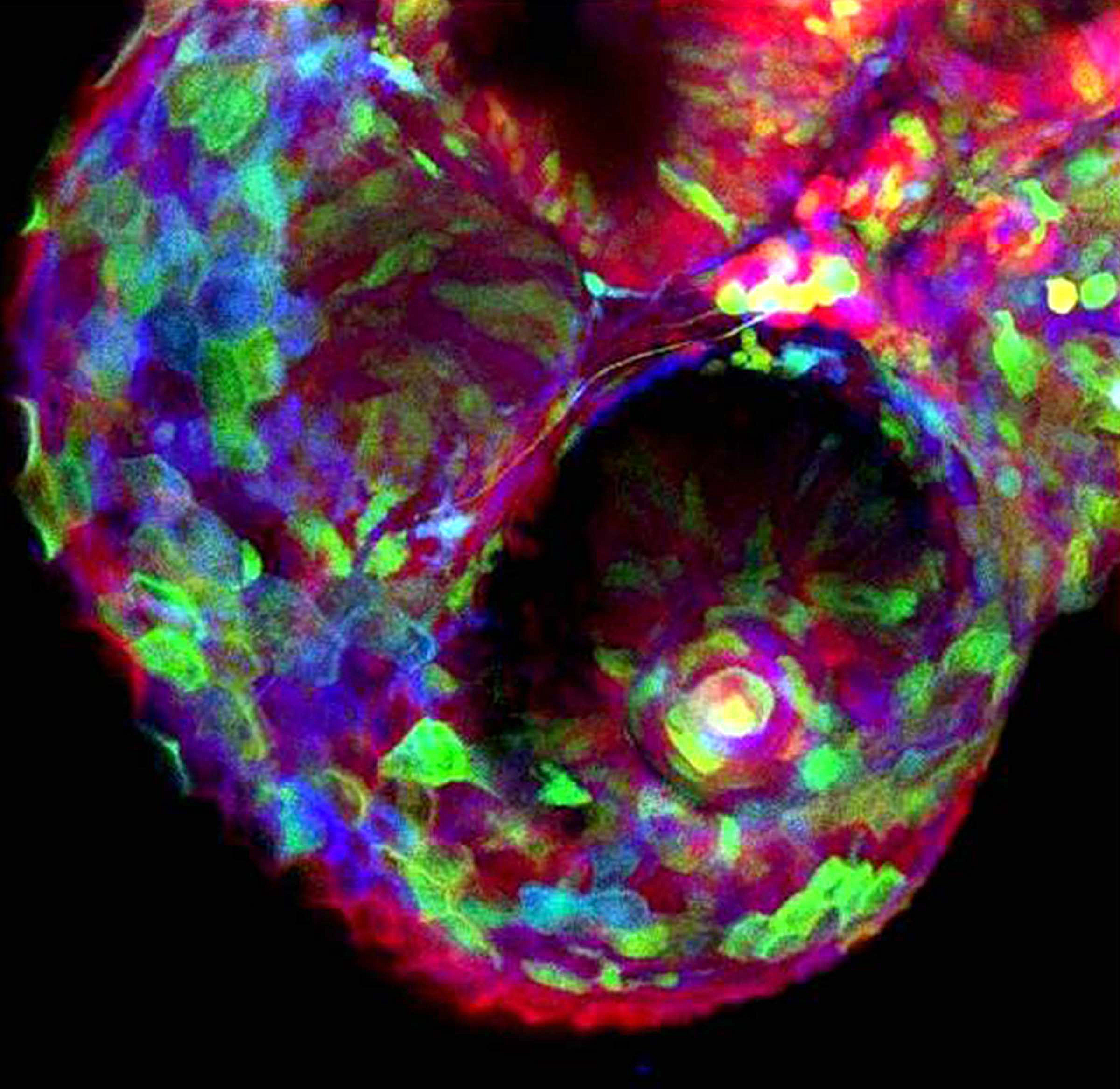
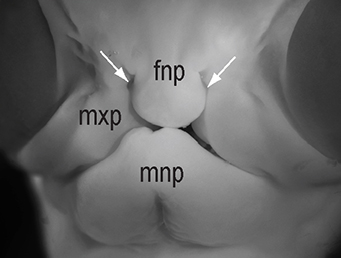 The chicken talpid2 and talpid3 mutants display a range of developmental phenotypes including craniofacial and limb defects. Although links to the sonic hedgehog (SHH) pathway had been proposed, the molecular nature of these mutations remained unclear for many years. The talpid3 phenotype is known to be caused by mutation in a ciliary protein – consistent with the known function of the cilium in SHH signal transduction. Now (
The chicken talpid2 and talpid3 mutants display a range of developmental phenotypes including craniofacial and limb defects. Although links to the sonic hedgehog (SHH) pathway had been proposed, the molecular nature of these mutations remained unclear for many years. The talpid3 phenotype is known to be caused by mutation in a ciliary protein – consistent with the known function of the cilium in SHH signal transduction. Now (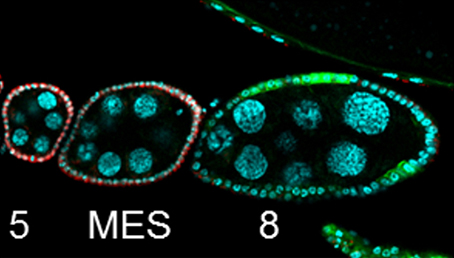 Organ growth and developmental progression must be coordinated with nutritional status. On
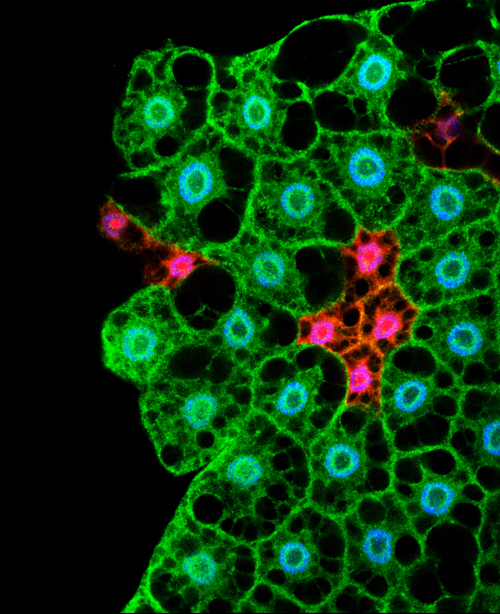
Organ growth and developmental progression must be coordinated with nutritional status. On 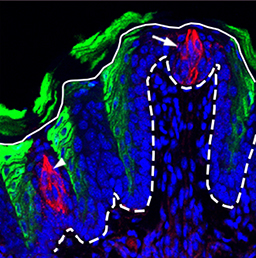 In the adult tongue, taste buds are located on taste papillae and are constantly renewed throughout life to maintain gustatory sensing. The sonic hedgehog (SHH) pathway has been shown to regulate taste bud formation in development, whereby SHH activity inhibits taste placode formation. Linda Barlow and colleagues now find (
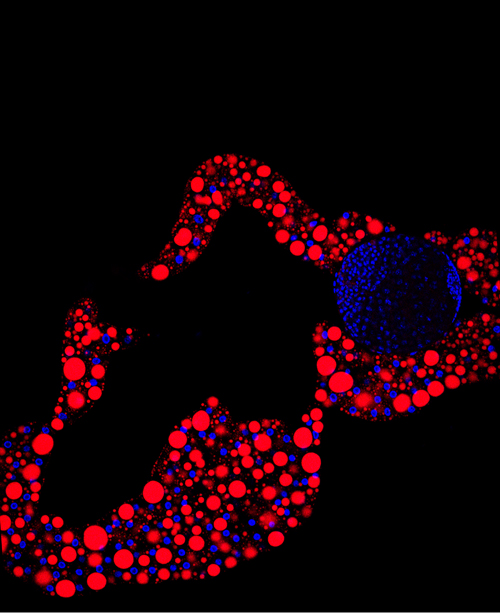
In the adult tongue, taste buds are located on taste papillae and are constantly renewed throughout life to maintain gustatory sensing. The sonic hedgehog (SHH) pathway has been shown to regulate taste bud formation in development, whereby SHH activity inhibits taste placode formation. Linda Barlow and colleagues now find (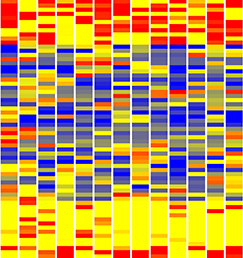 Single cell profiling technology now allows us to gain unprecedented insight into the complexities of gene expression within a developing tissue at the single cell level. Here (
Single cell profiling technology now allows us to gain unprecedented insight into the complexities of gene expression within a developing tissue at the single cell level. Here (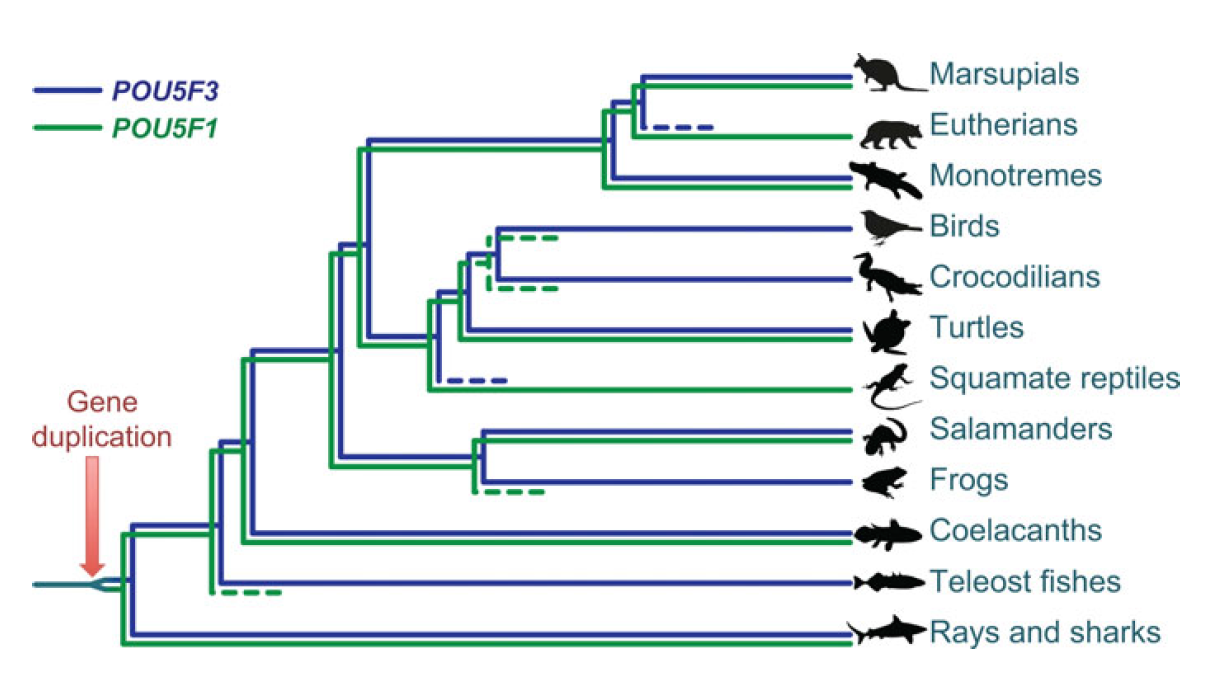 POU5F1 (OCT4) is a key regulator of stem cell fate, with homologues present throughout vertebrates. Frankenberg, Brickman and colleagues clarify the relationship between these homologues, aiming to resolve the confusion over the identity of the zebrafish gene. See the Spotlight on p.
POU5F1 (OCT4) is a key regulator of stem cell fate, with homologues present throughout vertebrates. Frankenberg, Brickman and colleagues clarify the relationship between these homologues, aiming to resolve the confusion over the identity of the zebrafish gene. See the Spotlight on p. Plants are able to adjust their growth in response to environmental changes, and this depends in part on their ability to establish polar protein distributions. Luschnig and Vert discuss the mechanisms involved in this process, focusing on plasma membrane proteins such as PINs. See the Review on p.
Plants are able to adjust their growth in response to environmental changes, and this depends in part on their ability to establish polar protein distributions. Luschnig and Vert discuss the mechanisms involved in this process, focusing on plasma membrane proteins such as PINs. See the Review on p. Developmental biologist Julian Lewis sadly passed away last April. Paul Martin and David Ish-Horowicz look back on his life and work. Read on p.
Developmental biologist Julian Lewis sadly passed away last April. Paul Martin and David Ish-Horowicz look back on his life and work. Read on p.