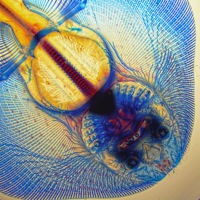
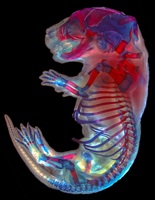
A horse is a horse of course of course…
Posted by Kim Cooper, on 24 April 2012
Unless it’s a Przewalski’s horse!
Oh where to even start! The embryo collections came to completion on Wednesday, and now we are on to Phase II. We returned to Urumqi on Thursday so I could give a talk at Xinjiang Normal University and we could pick up Talia at the airport on Friday. Sarah and I decided to take the public bus back to the city since we weren’t carrying much and were feeling adventurous. That involves taking a local bus for about 50 cents from 222 to Fukang and then the long distance bus from Fukang to Urumqi for about $2. By the way, in past years I thought the town near the field station and the city 20 minutes away were both Fukang. I always called one Fukang town and the other Fukang city. But this year I finally learned that the agricultural community here near the field station is “Regiment 222”. There is a long back history that I will leave you to explore and question for yourself.
Upon returning to Urumqi, Sarah and I met up with our friend from one of the labs here who is delightful. She spent a couple of years in Reno as a postdoc, so she’s familiar with American culture and personalities. Sarah wanted to introduce me to Pizza Hut in China, which is a fancy date destination. We did a sampling of the menu and stayed until close – cackling so much we probably interfered with a few romantic evenings. It started because I had been waiting on the bus while Sarah went to the market in 222 to get water and yogurt for the trip. To entertain myself, I was looking up the word for “wrench” that I had learned from a student at the field station when I wanted to take the regulator off our CO2 tank. Chinese is made up of a lot of compound words, so the word for wrench is ban (to pull) shou (manually). I have a dictionary on my phone that gives the meaning for each character and other compound words that use that character, so I accidentally discovered another compound word using the character shou that has a sexual meaning. I had shared this discovery with Sarah who not so innocently brought it up to our friend in the way of “I learned a new word”, and she intentionally pronounced it with the incorrect tone so that our friend would puzzle it together to figure out what she was asking. Her face exploded in shock, and she laughed. So that started a whole conversation about the worst words in English and Chinese, and I realized that learning a bad word as an adult doesn’t give it the same meaning. She could throw around some of the worst words in English as though she was saying “table” and “chair”, but when we asked her to tell us bad words in Chinese it was like we were asking her to stab her own hand. Likewise, we could toss around those words playfully in Chinese, but there is no visceral meaning. It’s as if you have to have had your mother threatening to wash your mouth out with soap to really feel the wrongness of a word.
On Friday morning, I was picked up and delivered to Xinjiang Normal University to give a talk in the biological sciences department on invitation from my hosts at the arrival banquet. They are all really friendly and enthusiastic to have me here. After my talk and tour of the natural history museum (great teaching resources), we walked across from the campus to a restaurant for another lunch time banquet. This was more informal than the baijiu fest from the first night, but again the fish hit the table and bottles of pijiu (beer) were opened. Speeches all around. The dean of the college of life sciences couldn’t make it to my talk but made an effort to come for lunch even though she was only able to join for the last half. She said she likes me. I joked that it’s because she knows I can drink well. She’s great. We talked a little about careers as women, and I asked about the numbers of women who start careers in biology at the masters level compared to the number of women who achieve full professorship. The Normal universities are teacher’s schools, so there is a slightly higher percentage at her institution, but it still hovers around 20% even though greater than 50% start out at the lower levels. I knew what the answer would be but asked “Why?” anyway. Of course she said, “Because they want to devote their time to family.” I asked if she thought it would ever be possible in China for men and women to both contribute equally to caring for the family so that both could have fulfilling careers if they want, and she said “No” (even though she later said that she is married and has a son, and her husband is very supportive which is what allowed her to reach the position of dean). One of the young men on the faculty who is recently married loudly protested and started arguing with her that yes, men will take on a more equal role and support their wives. I didn’t understand the rest of the argument since it was all in Chinese, but a young woman across the table from me who is a new professor and has a 15 month old son of her own just stared at me with amazement at what I’d started. I just gave a little grin and a wink to let her know I knew exactly what had happened and watched the debate unfold.
Later that evening, Talia arrived from Boston, and I managed to successfully retrieve her from the airport even though I hadn’t written down any of her flight info, didn’t know her airline, and didn’t know what terminal she was flying into. Thank goodness for small airports. I’m terrible about doing things like this. We kept her up the first night and planned outings for Saturday to get her over the jetlag. Sarah knew of a vegan restaurant she wanted us to try since Talia is vegetarian, and Xinjiang is the Chinese equivalent of Texas. We had several of the common Chinese dishes, except that everything that looked and tasted a lot like meat had no animal components at all. Really interesting. We then met up with my Uygur friend to go south to where I’ve lived before, because I wanted to show Talia all my favorite places and let her and Sarah get to know my dear friend. We got out of the cab, and it was an immediate sensory overload. The food vendors were starting to set up their carts in the area of the night market, so we wound our way through the rows of tumeric dusted roast chickens, sheep’s heads, and mounds of glass noodles. Talia and Sarah decided to get some sort of frighteningly fake sweet beverage because it was a shooting fountain of neon orange. We darted into a Uygur medicine shop where a man read our health histories in our pulses. I think he was a bit of a quack though – I’ve done this before and felt like the guy who saw me was at least paying attention to my skin tone and the health of my fingernails. This guy seemed a little cocky, barely looked at me, and his major comment was that I don’t absorb enough nutrition from my food. Not a startling discovery.
The narrow side streets are bustling with foot traffic, so we inserted ourselves in the river and wandered about watching as all the locals did their evening shopping. The tourist markets were all closing, but they mostly sell a bunch of kitschy things that aren’t made in this area anyway. It’s far more fun to roam the streets with the guys selling t-shirts and shoes shouting “Besh quai, besh quai, besh quai!!!” which means 5 RMB in Uygur. I made one guy laugh by joining in his call and smiling to let him know I wasn’t making fun of him. We found ourselves passing a halal butcher right after they had decapitated a lamb. It hadn’t yet been eviscerated, but the pelt lay in a pile on the ground, the head and feet were no more, and it was hanging by a wire cable threaded through the Achilles’ tendons over a bucket of fresh blood. Had we only passed by 10 minutes earlier, we would have gained a new appreciation for where our food comes from.
Wandering these streets again with my friend brought back a flood of memories from past years, as he and I reminisced shops we’d been to, funny things we’d seen, dinners we’d had. It was nice to share memories and tell Sarah and Talia our stories while watching them write stories of their own. And I keep learning more and more about Islam and life in this part of the world that hurts my heart and brain to think about how little we as Americans really know about this great big world past our borders. After yet another fantastic meal and amazing conversation, the three of us girls left my friend behind since that neighborhood is his home and hopped back into a taxi to return north to the institute. Once we got to the hotel, I kept Talia up another hour sharing my experiences and observations of the ethnic culture clashes of this region, and right as we were drifting off to sleep at a quarter to 2 am, I got a text message to my Chinese cell phone. It was a professor at the institute who wanted to be the one to bring us back to the field station asking what time he should meet us. Seriously. He sent me a message at 2 am. Sarah had warned me about this. The phone culture is strange. They can call or text at any time from anywhere, and it’s all okay. So I replied politely, said I was going to sleep, and turned off my phone.
This morning after some lengthy discussions and slight political issues, we arranged for him to pick us up since he really wanted to take us to the Przewalski’s horse breeding center. That was worth the 2 am text message and ensuing drama. And worth braving the dust storm. Somehow overnight the temperature dropped about 40 degrees, and the winds came howling in from Russia. If I wasn’t Russian before, I am now on the inside and out. I feel like my eyeballs, my skin, and the insides of my nose and ears are coated with the dust carried from the winds of the north. But it was worth almost being lifted off my feet to see these amazing horses. Apparently before the breeding center started in 1985, there were only about 2,000 Przewalski’s horses in the wild. They are a species of wild horse that is native to Xinjiang, Inner Mongolia, and Mongolia (Inner Mongolia is a province of China. Mongolia is a country.) The breeding center started with 18 horses and now has a population of about 400. They have been reintroducing many of the captive bred horses into the wild, and have managed to have a real positive impact on the population of this species. They are truly wild horses. They are smaller than what you think of as a horse and look more like a donkey or a mule with a short mane and short flat tail. They still have most of their thick winter coat, and the younger horses look like they are wearing leg warmers. And they are mean! They get really grumpy and fight with each other. They have to be kept in families of one male and about 5-6 breeding females. Even the females will fight with each other. When one gets irritable, she backs her rump into another and just keeps pushing and pushing until the other gets annoyed. Then they separate just enough to get a good kick it. But it isn’t just a kick. It’s an ears back, teeth bared, two-footed bucking kick. Sarah kept making noise to spook and separate them, but that’s just what they do. They’re really truly wild horses.
The rest of the drive was fascinating. All along the mountains is mining country, so there are great big holes in the earth surrounded by digging equipment. And even though it isn’t a major oil field like what we wandered into up north, there are pumpjacks dotting the landscape. In addition to that, it’s a major industrial zone, so one after another we passed by some sort of manufacturing/mining/power station/etc type of location. The air was thickly brown with the dust, but not just from the dust. All around are mounds of coal piled high, and the dust from the coal gets picked up in the wind right along with the barren earth and sand. So it’s no wonder when I scratch my face, the underside of my nails is black. In fact, I have been writing this while waiting for my hot water heater to warm up. I think that ought to be done by now, so I’m going to shower my nasty self and get to bed.


 (4 votes)
(4 votes) (7 votes)
(7 votes)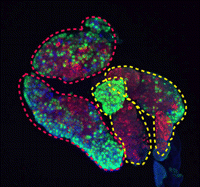 During development and homeostasis, it is essential to coordinate growth with the availability of nutrients. The interconnected insulin/IGF (IIS) and target of rapamycin (TOR) pathways integrate tissue growth with dietary conditions in Drosophila, and now Marc Haenlin and co-workers show that these pathways play a crucial role during haematopoiesis in the Drosophila lymph gland (
During development and homeostasis, it is essential to coordinate growth with the availability of nutrients. The interconnected insulin/IGF (IIS) and target of rapamycin (TOR) pathways integrate tissue growth with dietary conditions in Drosophila, and now Marc Haenlin and co-workers show that these pathways play a crucial role during haematopoiesis in the Drosophila lymph gland (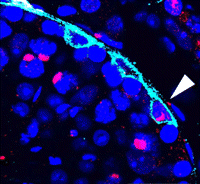 Spermatogonial stem cells (SSCs) have the remarkable ability to self-renew and support spermatogenesis throughout life. It is known that fibroblast growth factor 2 (FGF2) promotes SSC self-renewal but the factors acting downstream of FGF2 are unknown. Here, Takashi Shinohara and colleagues show that FGF2 regulates SSC self-renewal via MAP2K1 and the Etv5 and Bcl6b genes (
Spermatogonial stem cells (SSCs) have the remarkable ability to self-renew and support spermatogenesis throughout life. It is known that fibroblast growth factor 2 (FGF2) promotes SSC self-renewal but the factors acting downstream of FGF2 are unknown. Here, Takashi Shinohara and colleagues show that FGF2 regulates SSC self-renewal via MAP2K1 and the Etv5 and Bcl6b genes (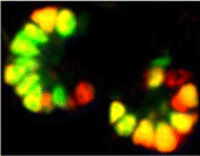 In the developing pancreas, the branched epithelium can be separated into tip and trunk regions, with the tip domain generating acinar cells, and the trunk domain differentiating to endocrine and duct fates. Although Notch signalling is known to be important for proper pancreatic development, particularly in maintaining the progenitor state and inhibiting premature endocrine differentiation, its precise roles in regulating cell fate remain unclear. Here (
In the developing pancreas, the branched epithelium can be separated into tip and trunk regions, with the tip domain generating acinar cells, and the trunk domain differentiating to endocrine and duct fates. Although Notch signalling is known to be important for proper pancreatic development, particularly in maintaining the progenitor state and inhibiting premature endocrine differentiation, its precise roles in regulating cell fate remain unclear. Here (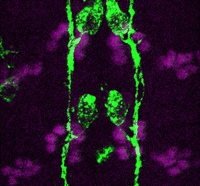 Accurate axonal pathfinding relies on the tightly regulated expression of guidance cues and their receptors, but the links between transcriptional regulators and downstream guidance factors are poorly understood. Genetically amenable Drosophila motoneurons provide an ideal system for analysing the control of guidance receptor expression. It is known that two transcription factors, Even-skipped (Eve) and Grain (Grn) are expressed in the aCC and RP2 motoneurons, and that projection of these neurons to the muscle requires the Netrin receptor Unc-5. Now, Juan-Pablo Labrador and colleagues dissect out the relationships between these factors (
Accurate axonal pathfinding relies on the tightly regulated expression of guidance cues and their receptors, but the links between transcriptional regulators and downstream guidance factors are poorly understood. Genetically amenable Drosophila motoneurons provide an ideal system for analysing the control of guidance receptor expression. It is known that two transcription factors, Even-skipped (Eve) and Grain (Grn) are expressed in the aCC and RP2 motoneurons, and that projection of these neurons to the muscle requires the Netrin receptor Unc-5. Now, Juan-Pablo Labrador and colleagues dissect out the relationships between these factors (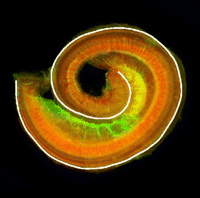 The atypical cadherin Fat (Ft) is crucial for planar cell polarity (PCP) in Drosophila. Four ft homologs (Fat1 to Fat4) have been identified in mammals, but the functional roles of these homologs and any possible redundancies between them are unclear. Here, Helen McNeill and colleagues study the genetic interactions between mammalian Fat genes and show that Fat proteins act both synergistically and antagonistically to regulate multiple aspects of tissue morphogenesis in mice (
The atypical cadherin Fat (Ft) is crucial for planar cell polarity (PCP) in Drosophila. Four ft homologs (Fat1 to Fat4) have been identified in mammals, but the functional roles of these homologs and any possible redundancies between them are unclear. Here, Helen McNeill and colleagues study the genetic interactions between mammalian Fat genes and show that Fat proteins act both synergistically and antagonistically to regulate multiple aspects of tissue morphogenesis in mice (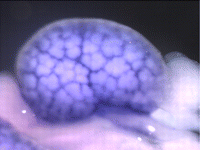 The kidney comprises multiple cell types of both epithelial and mesenchymal origin, with highly defined regional subdivisions in the ductal systems. A full understanding of kidney development requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney (
The kidney comprises multiple cell types of both epithelial and mesenchymal origin, with highly defined regional subdivisions in the ductal systems. A full understanding of kidney development requires that each cell type can be uniquely identified by specific molecular markers. To this end, Andrew McMahon and colleagues have undertaken a comprehensive analysis of the RNA expression patterns of nearly one-thousand transcription factors in the embryonic mouse kidney ( (No Ratings Yet)
(No Ratings Yet)