After a heart attack, heart muscle is irreparably damaged, but a paper in Nature now reports that adult mouse hearts have a source of progenitor cells that can form new muscle cells after heart injury.
A few years ago, studies showed that embryonic epicardial progenitor cells contribute to the cardiomyocyte lineage in developing mouse hearts. These cells were marked by the expression of a key embryonic epicardial gene, Wt1, but Wt1 is not expressed in adult tissues.
The group of Paul Riley at UCL now reactivated Wt1 expression in adult mouse hearts by priming them with thymosin β4 (Tβ4) and inducing injury. This pointed to an adult pool of progenitor cells, marked by Wt1, which could form new cardiomyocytes after myocardial infarction. What’s more, this process was upregulated in response to Tβ4. A few years ago, Riley’s group already showed that Tβ4 also induces formation of blood vessels from epicardial progenitors.
In a video interview with The Scientist, Riley summarized his paper, and emphasized how they built upon previous studies in embryonic heart development to find this new source of adult myocardial progenitors.
Repairing hearts from thescientistllc on Vimeo.
“The key point for us has always been moving back to embryonic development and identifying cells that are key to formation of the organ, that would then translate to repair in the adult.” – Paul Riley (from the interview above)
How exactly Tβ4 induces increased Wt1 expression and cardiomyocyte formation isn’t yet known, but could this be a new therapeutic for heart attack patients? Unfortunately, Tβ4 is not the most practical drug. It would need to be administered before a heart attack, so could only be used as a preventive measure for people who already know they’re at risk, and it’s not available as a pill – only as injection. But a big step toward any form of therapy would be to find out how Tβ4 works at a molecular level to differentiate the progenitor cells to cardiomyocytes upon injury, and, as Riley mentions in the video above, that will be the next step in their research.
update: F1000/The Scientist have some more videos from this lab on their blog.
(more…)
 (1 votes)
(1 votes)
 Loading...
Loading...
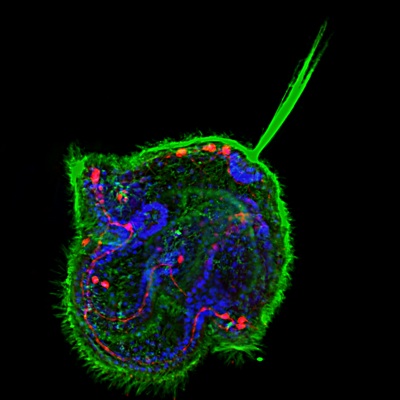 Pilidium larva of the Nermertean, Cerebratulus lacteus. Acetylated tubulin (green), serotonin (red), nuclei (blue, DAPI).
Pilidium larva of the Nermertean, Cerebratulus lacteus. Acetylated tubulin (green), serotonin (red), nuclei (blue, DAPI).

 (4 votes)
(4 votes) (2 votes)
(2 votes)