BSDB Gurdon Studentship Report – Aleksandr Markov
Posted by Alexander Markov, on 5 January 2023
Role of PAX6 in human cerebral organoid development
The cerebral cortex contains two major classes of neurons: excitatory and inhibitory. The imbalance between them is postulated to underlie some autism spectrum disorders (ASDs) (Rubenstein, 2010; Uzunova et al., 2016). The transcription factor PAX6 is an important regulator of embryonic forebrain development and mutations in PAX6 have been associated with ASDs (Kikkawa et al., 2019; Manuel et al., 2015). It is therefore possible that PAX6 could be involved in the regulation of the inhibitory-excitatory balance, hence playing a role in the development of ASDs. Supporting this hypothesis, recent findings indicate that PAX6 deletion in mice leads to the appearance of ectopic GABAergic (inhibitory) cells (Manuel et al., 2022). While animal data are useful, the mechanisms of this effect, and whether it is present in humans remains unclear, as human neurodevelopment is difficult to study directly. However, human cerebral organoids offer an exciting, new way to directly investigate early human embryonic neurodevelopment, including the effects of PAX6 mutations (Mason & Price, 2016). Hence, as a first step towards uncovering the mechanisms of the possible PAX6-dependent regulation of the inhibitory-excitatory balance in humans, this project aimed to examine PAX6-/- organoids at early developmental stages to investigate the effects of PAX6 mutations.
Methods
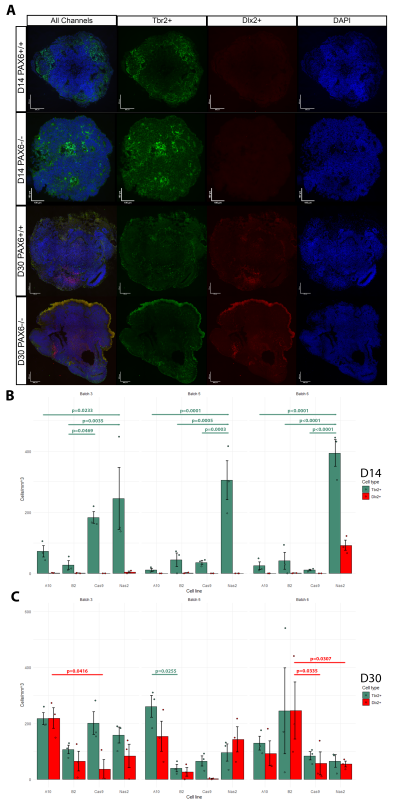
The Mason Lab has previously grown cerebral organoids from PAX6-/- induced pluripotent stem cells and PAX6+/+ controls. At culture day (D)60, the cellular phenotypes of these organoids were tested using scRNA-seq analysis. Indeed, ectopic GABAergic populations were observed in the mutants and not in controls. To examine the earlier stages of PAX6-/- organoid development, I analysed three batches of organoids originating from four cell lines (Controls: Nas2,Cas9; Mutants: A10,B2) at D14 and D30. Organoids were cut into 10μm sections. Inhibitory/excitatory neurons were visualized by tagging Tbr2 (marker of excitatory glutamatergic neurons/progenitors) and Dlx2 (marker of inhibitory GABAergic neurons/progenitors) with fluorescent antibodies. Hence, two distinct cell types could be visualized using fluorescent microscopy (Fig.1A). The number of Tbr2+ and Dlx2+ cells was counted in three randomly selected sections from each organoid using ImageJ software. The cell count was normalized by area of each section and averaged per organoid (Fig.1B-C).
Results
No D14 organoid (apart from an anomalous batch 6 Nas2 organoids) featured Dlx2+ cells. Hence, ectopic GABAergic cells likely arise after D14, which narrows the window of investigation in potential future research. In many cases, the number of Tbr2+ cells was significantly higher in the Nas2 organoids when compared to other cell lines, including the second control line, Cas9 (Fig.1B). This may suggest the presence of unknown variables which affected organoid development, or possibly that PAX6 has an effect on the speed/timing of Tbr2+ neuron emergence.
Both mutant and control organoids at D30 exhibited comparable counts of Tbr2+ and Dlx2+ cells, with significant differences in cell counts present between only a few groups (Fig.1C). This is surprising, as prior literature suggests that ectopic GABAergic cells (in PAX6-/- mice) appear at some point during development due to environmental signals such as Shh found in cerebrospinal fluid (CSF) and/or immigrating interneurons (Manuel et al., 2022). It seems that in human cerebral organoids (in the absence of CSF or immigrating interneurons) GABAergic cells emerge as a normal part of development whether PAX6 is present or not, but disappear by D60 in controls.
PAX6 could possibly be triggering the death of Dlx2+ cells between D30 and D60. PAX6 could also be causing Dlx2+ cells to switch to an excitatory fate by D60. It would be interesting to expand upon these findings by conducting lineage tracing experiments: this would allow for the tracking of the changes in cellular phenotypes to reveal whether the presence of PAX6 affects cell fate decision making.
My experience at Mason Lab
I found this project very exciting. The problem-solving aspect of research following inevitable experimental failures was genuinely great. It was very interesting to brainstorm the next steps when it becomes apparent that the current experimental procedure is not yielding any results. For instance, originally the experiment involved tagging dividing cells with EdU at D13.5, to see whether these cells switch between Tbr2+ and Dlx2+ identities between D14 and D30. However, the experiment had to be modified as the EdU fluorescent signal became too faint at D30 due to dilution during cell division.
The project also showed me the more labour-intensive side of research as I spent many days cutting organoid sections at the cryostat. After the baptism of fire by cutting my finger, I eventually found it a compelling task. As one of the Mason Lab members said, “it builds character”. I was also introduced to fluorescent microscopy and image analysis using ImageJ. The former forced me to become organised and efficient, as one must tame the unruly microscope and take the hundreds of pictures before the booked time runs out. The latter I found challenging as it was my first time using image analysis software, but it will undoubtedly be useful in my future work.
Every part of this project, no matter how menial or monotonous it might have seemed to an observer, I found thrilling. I have obtained experience in numerous laboratory techniques, and again reaffirmed research as my chosen career path. I am very grateful to Mason Lab and BSDB for this opportunity, and I am looking forward to being able to conduct projects of larger scales in the future.



 (2 votes)
(2 votes) (No Ratings Yet)
(No Ratings Yet)