Autonomous traffic – Wnt cytonemes lead the way.
Posted by Steffen Scholpp, on 2 October 2018
by Lauren Porter and Steffen Scholpp
Living Systems Institute, University of Exeter, UK
The importance of Wnt signalling in developmental processes, wound healing and stem cell control has long been established. Historically, scientists attributed the transport of Wnt proteins from the source to the receiver cell to simple diffusion, however, this explanation did not seem sufficient to support the precise delivery of Wnt proteins required to satisfy healthy development. In 2015, our group discovered that finger-like membrane protrusions termed cytonemes delivered the Wnt proteins to responsive cells, but the governance of Wnt-positive cytonemes was yet to be elucidated. A new paper published by our group indicates that Wnt retains sovereignty over its own delivery by binding to the kinase Ror2, which acts upstream of cytoskeletal regulators to manage the formation of Wnt-positive cytonemes.
Wnt signalling is absolutely fundamental to embryogenesis. Present in all metazoans, Wnt proteins have been closely examined in many model organisms such as the fruit fly, mouse, frog, and zebrafish. Since its discovery in mice by Roel Nusse and Harold Varmus, and in parallel by Christiane Nüsslein-Volhard and Eric Wieshaus in Drosophila, it made its début as the viral insertion site int-1 and segment polarity gene Wingless combined as Wnt-1 in the eighties. In the three decades since it was unearthed, Wnt has gained traction as a family of proteins with famously important roles in embryonic development, cell survival, proliferation and stem cell regulation, which act by the formation of concentration gradients over responsive cell types.
Notwithstanding the accumulation of knowledge concerning the action of Wnt proteins, exactly how Wnt protein gradients are secreted in a controlled manner is a source of debate; several theories persist, including simple diffusion and exocytosis. Despite the appealing adherence to the principles of Occam’s Razor, it seemed that a more complex mechanism was required to explain the precision of Wnt delivery. Indeed, in 2015, our group discovered such a mechanism in the zebrafish, Danio rerio. Wnt proteins were in fact delivered from the sender cell by finger-like membrane protrusions carrying Wnt proteins at their tips, allowing full end-end control of Wnt morphogenetic signalling by the sender cell. These membrane protrusions, termed cytonemes, had earlier been suggested to carry the Dpp and Hedgehog signalling molecules in Drosophila development. Now, the function of cytonemes in the facilitation of developmental processes has been expanded to the delivery of Wnt signals in vertebrates.
Cytonemes mobilize Wnt protein
Membrane protrusions are not unusual and are commonly referred to as filopodia. The distinction between cytonemes and filopodia is the cargo transported – the signalling molecules. Both, however, are actin-dependant and, we discovered, contingent on Cdc42, a small Rho family GTPase integral to dynamic F-actin assembly when we were based at the Karlsruhe Institute of Technology (KIT) still. Manipulating the length and number of Wnt-positive cytonemes led to malformed embryos because of aberrant tissue patterning, and therefore we hypothesised that the regulation of formation, timing, number and length of Wnt-positive cytonemes must be tightly controlled in order to produce the fine balance of Wnt gradients that produce wild type tissue patterning.
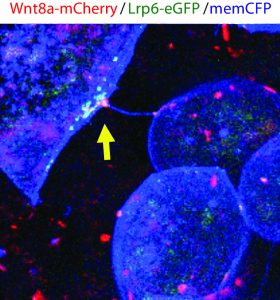
The method and importance of Wnt gradient formation thus elucidated, we set out to determine the upstream control of the genesis of Wnt-positive cytonemes. Benjamin Mattes, PhD student in the lab, decided to examine whether a kinase could in fact be a candidate due to their multifaceted roles in cellular processes in 2016. Preliminary findings indicated that his hunch was correct, and multiple kinases induced changes in the number of membrane protrusions. To narrow down the selection of kinases, we collaborated with computer scientists at KIT who used modelling techniques to select the tyrosine kinase receptor Ror2. Unlike the other kinases identified by the screen, Ror2 is involved in both Wnt signal transduction of the non-canonical Wnt signalling pathway and in regulation of the F-actin cytoskeleton, thus uniting two key molecular functions in the building of cytonemes.
Ror2 controls Wnt cytonemes in zebrafish
To truly hammer home the importance of Ror2, Benjamin decided to try some studies in both zebrafish embryos and in vitro in zebrafish PAC2 cell cultures. The result of Ror2 overexpression was increased number of Wnt-positive cytonemes, inferring that Ror2 was involved in the nucleation of cytonemes, and also that it worked upstream of small Rho GTPase Cdc42, which had earlier been implicated in the regulation of the actin cytoskeleton used to build the protrusions via the non-canonical Wnt signalling pathway. Surprisingly, filopodia which did not carry Wnt8a-GFP did not change in number. Consistently, when Ror2 was knocked down, there was a significant decrease in the number of cytonemes and again filopodia number was unaffected. Regulators of cytonemes presented so far do interfere also with filopodia. However, our data suggests that Ror2 is a cytoneme specific regulator – the first one at least to our knowledge.
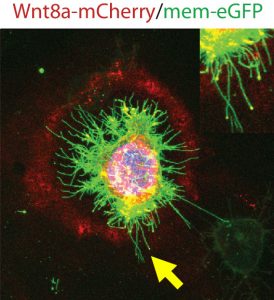
Subsequently, Benjamin set out to investigate how exactly Ror2 was able to stimulate cytoneme formation by using image-based approaches, which showed that Wnt family member Wnt8a – which has an established role in zebrafish embryonic development – and Ror2 associate and move together in a complex. Still, he could not entirely prove that Ror2 and Wnt8a bind each other – until he collaborated with the physicist Uli Nienhaus at the KIT who works on developing novel super-resolution techniques. Over one nail-biting summer, they worked together to finally prove that Ror2 and Wnt8a bind together in the living zebrafish embryo – in membrane associations with presumably other mystery players.
At the end of this summer, the lab moved from the KIT in Germany to the newly established Living Systems Institute (LSI) in Exeter, UK. Benjamin continued with the experiments in the world-class institute immediately. In endless imaging sessions using the newly purchased confocal microscope he found that these Wnt8a-Ror2 membrane associations activate PCP signalling in ligand-producing cells, which induces the formation of cytonemes ready to deliver their cargo – Wnt proteins. Therefore, it appears that Wnt proteins retain sovereignty over their own delivery by controlling the nucleation of cytonemes. The complex gradients of Wnt proteins that dictate tissue patterning are governed by Wnt itself through binding with Ror2, activation of PCP signalling, and subsequent cytoneme nucleation.

Wnt cytonemes in cancer tissue
The dependence of cytoneme nucleation on Ror2 signalling raises some interesting questions – for example, does Ror2 regulate Wnt signalling in other paracrine processes, such as cancer cell proliferation? Using a co-culture of AGS human gastric cancer cells, Benjamin found that Ror2 enhanced Wnt response in adjacent cells by greater nucleation of cytonemes and therefore increased cancer growth.
For some cells, paracrine Wnt signalling is required throughout their whole lifespan. To investigate whether Ror2 has a role in regulating cytoneme formation in these cells, we started a collaboration with mouse geneticist David Virshup of the Duke-NUS Medical School in Singapore. The intestinal crypt requires a constant supply of Wnt signalling, and David’s team demonstrated recently that the myofibroblasts that surround the crypt have an abundance of cytonemes. When we silenced Ror2 in myofibroblasts, they produced less cytonemes and the intestinal crypt cells that they surround die.
Here, we come to almost present day. All that remains to be said is that we are grateful that last summer, Christiane Nüsslein-Volhard visited the Living Systems Institute at Exeter as key note speaker for its official opening and remarked that she was very pleased to see that Wingless/Wnt trafficking is on its way to being finally solved, and that zebrafish are playing such a large part in that. This said, there is undoubtedly more to learn; cancer cell proliferation and the survival of the mouse intestinal crypt are just two examples where our insights into cytoneme nucleation and the Wnt signalling generated are proving to be instrumental. It seems that where Wnt signalling is concerned, it is in sickness and in health, to death us do part.

References
Mattes B, Dang Y, Greicius G, Kaufmann LT, Prunsche B, Rosenbauer J, Stegmaier J, Mikut R, Özbek S, Nienhaus GU, Schug A, Virshup DM, Scholpp S. Wnt/PCP controls spreading of Wnt/β-catenin signals by cytonemes in vertebrates. eLife. 2018 Jul 31;7. pii: e36953. doi: 10.7554/eLife.36953.
Stanganello E, Hagemann AI, Mattes B, Sinner C, Meyen D, Weber S, Schug A, Raz E, Scholpp S. Filopodia-based Wnt transport during vertebrate tissue patterning. Nat Commun. 2015 Jan 5;6:5846. doi: 10.1038/ncomms6846.


 (4 votes)
(4 votes) (No Ratings Yet)
(No Ratings Yet) from a recipient of a Development Travelling Fellowship, funded by our publisher The Company of Biologists.
from a recipient of a Development Travelling Fellowship, funded by our publisher The Company of Biologists.






 Barcelona, 26 September 2018.- Metastatic tumours are characterised by a high degree of chromosomal instability—that is to say a constant change in the number and structure of chromosomes they hold. In spite of this observation, it is unknown whether chromosomal instability contributes to the metastatic process.
Barcelona, 26 September 2018.- Metastatic tumours are characterised by a high degree of chromosomal instability—that is to say a constant change in the number and structure of chromosomes they hold. In spite of this observation, it is unknown whether chromosomal instability contributes to the metastatic process.