The people behind the papers – Kimberly McArthur & Joseph Fetcho
Posted by the Node Interviews, on 20 July 2017
Development depends on cells coming together in the right place to form functioning tissues. Our latest paper in this series was published recently in Current Biology and asks what happens to the structure and function of the nervous system when motor neuron migration – and hence final position – is disrupted. We caught up with postdoc Kimberly McArthur and her PI Joseph Fetcho, Professor at the Department of Neurobiology and Behaviour at Cornell University.
Joe, can you give us your scientific biography and the main questions your lab is trying to answer?
JF I was always curious about different types of animals, but I was particularly interested in snakes and how they moved. As a graduate student, I studied the locations of spinal motor pools across multiple species, and found that anamniotes (like snakes and fish) – unlike amniotes (like mammals) – lack a clear topographic relationship between motor neuron location in the spinal cord and muscle target location in the periphery. As a postdoc, I started working more with physiology and circuit structure in goldfish, studying the hindbrain Mauthner cell circuit that generates fast escapes, because I thought I could use it to figure out a circuit for a vertebrate behavior from sensory input to motor output before I died. Once I started my own lab, I switched to studying neural circuits in larval zebrafish – which had been an important developmental model for many years, but which also had incredible potential as a vertebrate circuit and behavioral model because of its relative simplicity, optical transparency, and genetic accessibility. Since then, my lab has continued to use zebrafish to study the principles by which neural circuits generate behavior, and how these principles inform and are informed by our understanding of circuit development and evolution.
And Kim, how did you come to join the Fetcho lab?
KM In my graduate work, I studied state-dependent changes in hindbrain sensorimotor circuits in pigeons – seeing how vestibular reflexes were modified by the transition into flight, and looking for the neural correlates. I was puzzled by the (apparent) lack of topography in the vestibular nuclei, how neighboring neurons can have completely different response properties – and I was a bit frustrated that I didn’t have the tools to easily nail down the identity of the neurons with a specific set of physiological properties. I came across a pair of papers from Joe’s lab (Kinkhabwala et al. 2011 and Koyama et al. 2011, both in PNAS) demonstrating the link between early neuronal topography and circuit recruitment for interneurons in the hindbrain, using larval zebrafish. I was impressed both by the scientific finding and by the way in which the model facilitates a direct line of investigation from genetic profile to cell morphology to physiology to behavior – all in an intact vertebrate.
How did you come to be interested in the relationship between cell positioning and function in the nervous system? Had this been experimentally tested before your work?
JF My predoctoral research dealt with the role of neuronal positioning in spinal motor circuits, and some of our current work demonstrated the correlation between hindbrain interneuron positioning and circuit recruitment. Previous studies have certainly addressed the importance of neuronal positioning; indeed, there is a great deal of evidence that positioning can be critical for proper circuit development. And it’s sensible for the developing nervous system to lean heavily on positioning as a strategy for targeting synaptic connections, as most neurons arise in specific locations and end up in stereotyped locations within a given species. We were interested in the importance of migration and hoped to get a better understanding of exactly what changes in a case where neuronal position is altered by a genetic block of migration in a population of cells that also changes position in evolution. We figured it would help us understand why migration and position seem to matter so much. We did not expect to find so much unaffected by a large position change.
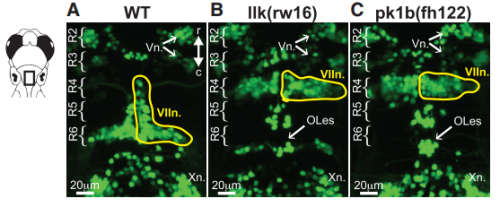
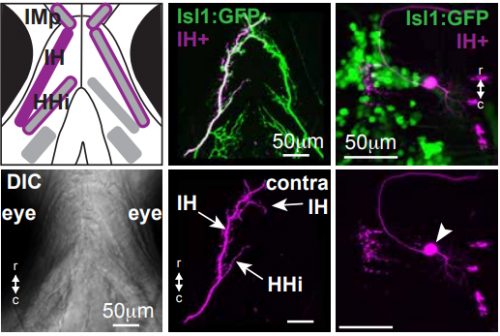
KM Other labs were using zebrafish facial motor neurons to study cellular mechanisms of neuronal migration, because these neurons execute an early caudal migration through the developing hindbrain. Those researchers had identified several mutations that spared the gross organization of the hindbrain but blocked the caudal migration of the facial motor neurons. We saw an opportunity to investigate the impact of this dramatic mis-positioning on the facial motor neurons themselves and the developing cranial motor circuits. We expected that there would be clear functional deficits in mis-positioned neurons, and we thought we’d be able to work out the details of how those deficits occur.
Can you give us the key results of the paper in a paragraph?
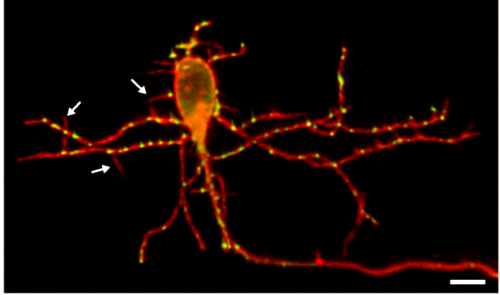
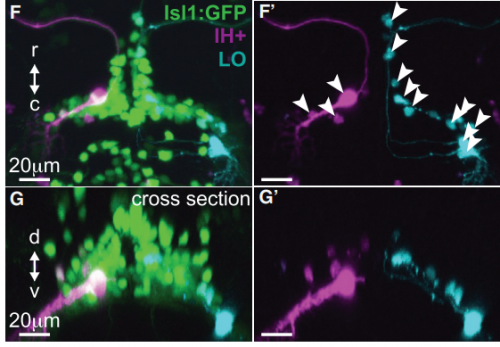
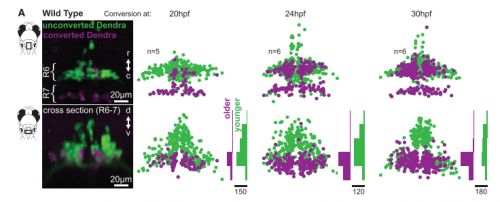
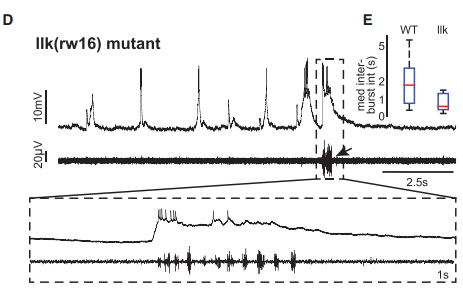
KM Contrary to our expectations, we found several lines of evidence indicating that facial motor neurons were robust to an abnormal shift in their segmental position. Even though their cell bodies and dendritic arbors were in entirely the wrong part of hindbrain, they still established intra-population topography (according to muscle target and relative age) and respiratory activity patterns (observed using intracellular electrophysiology and calcium imaging) that were very similar to wild type neurons. Indeed, wild type and mutant larvae exhibited similar respiratory behavior, which involves significant contributions from facial-innervated structures. Taken together, these results indicate that hindbrain motor networks can be surprisingly robust to a change in neuronal positioning – though there may be underlying differences in the motor circuits that we have yet to identify (which we’re pursuing now).
Is the resilience to change in position you’ve identified in motor neurons likely to be specific to these cells or a general feature of the nervous system?
KM I don’t believe that resilience to abnormal positioning is specific to facial motor neurons, but we also already know that it isn’t a general feature of every neuronal population – given previous work demonstrating that scrambling cell position can interfere with synaptic targeting in spinal cord, for example. Instead, I think that the developing nervous system deploys different synaptic targeting strategies in different circuits, and that some of these strategies are less sensitive to position than others.
Further, in the case of the facial motor neurons, I’m not convinced that positioning isn’t important. Facial motor neurons are born in roughly the same segmental location across vertebrates, but they end up in different places – suggesting that one location might be more adaptive than another in a particular species, depending on the dominant inputs to the facial motor neurons. It could be something to do with the metabolic cost of maintaining neural processes (i.e. wiring minimization), but I suspect that shifting segmental location might also affect the probability of synaptic input from a specific source (rather than specifying it in a deterministic fashion).
Finally, it’s interesting to note that facial motor neurons did establish the same cross-sectional topography in wild type and mutant larvae. Perhaps relative dorsoventral or mediolateral positioning is more critical than segmental (rostrocaudal) positioning for synaptic targeting in this specific population. This could be because many tracts in the hindbrain run longitudinally, so moving along that axis might have less impact on the inputs than moving in another direction.
And what might be the evolutionary implications of your findings?
KM I think of the developing nervous system as (implicitly) giving each neuron a set of instructions for locating potentially useful synaptic partners. Those instructions can be written using different strategies. For any given synapse, multiple strategies might work equally well – but if there is some perturbation (due to, for example, a genetic mutation or an abrupt environmental change), different strategies will fail in different ways. This will be reflected in how specific circuits respond to developmental perturbations (i.e. which perturbations actually cause dysfunction and disease), and it will shape the landscape of inter-individual variation that is available as a substrate for natural selection. Facial motor neurons, for example, have shifted their segmental location across phylogeny. Perhaps that’s possible because they are targeted for synaptic contact in such a way that core behaviors (like respiration) are robust to a shift in segmental position. This might give the facial nucleus a chance to sample (via mutation) novel, potentially adaptive synaptic connectivity without losing access to its original network.
When doing the research, did you have any particular result or eureka moment that has stuck with you?
KM I was initially a bit disappointed when I couldn’t find a clear functional difference between wild type and mutant facial motor neurons. I’d been framing my research as a case study in which to explore how abnormal positioning does impact function, in a particularly helpful animal model for working through the circuit-level implications. However, as I did more thinking and more reading, I realized how interesting and important this result might be. As useful as it can be to look for factors that (when disrupted) break the system, I think it’s also important to understand how much can go wrong without breaking the system. Indeed, I think this may be part of the reason that many human neurological disorders are so genetically complex: because breaking a single factor (or even a set of factors) involved in circuit formation and function doesn’t necessarily cause observable disease in every individual.
And what about the flipside: any moments of frustration or despair?
KM The biggest technical challenge for me was learning how to do whole-cell recordings in the larval zebrafish brain. When I was a graduate student, I learned how to do extracellular in vivo electrophysiology – which isn’t trivial, but you can start getting data relatively early if you get lucky with your electrode placement. When I started my postdoc, I spent several months learning how to handle the tools, do the dissection, work the scope, and finally record from cells. And I spent almost two months straight focused entirely on recording without actually getting any data. So, it was challenging to spend the first part of my postdoc mostly failing all day – but I also kinda like those moments, because it’s rewarding to overcome a challenge. And it worked out well in the end.
What are your career plans following this work?
KM I’m on the academic job market this year, applying for tenure-track faculty positions that combine research and teaching. I’d like to join a department where I can continue studying the development and function of neural circuits using larval zebrafish.
And what’s next for the Fetcho lab?
JF Recently, my lab has started to move into collaborations to advance technologies for neural circuit research, especially for looking with high resolution into brain and spinal cord of intact living adult zebrafish. The goal is to be able to study circuits and behavior in a vertebrate (even in one individual!) from embryo to adult.
Finally, what do you two like to do in NY state when you are not in the lab?
JF Not much. Lab has been my passion and my job. I mostly work, read, and think. I sometimes hang out in or near the woods behind our house looking for snakes and other animals including bears, pileated woodpeckers, turkeys, foxes and bobcats. Animals are my thing. People, not so much.
KM I think I’m your quintessential nerd! I get excited about going to the public library and checking out a stack of books. (No, seriously.) I try to balance my literary diet: nonfiction books about current events, plus lots of horror novels.
Kimberly L. McArthur, Joseph R. Fetcho. 2017. Key Features of Structural and Functional Organization of Zebrafish Facial Motor Neurons Are Resilient to Disruption of Neuronal Migration. Current Biology. 27 (12)1746–1756
This is the 23rd episode of our People Behind the Papers series; the full archive can be viewed here


 (No Ratings Yet)
(No Ratings Yet)


 Research into human development involves the use of human embryos and their derivative cells and tissues. How religions view the human embryo depends on beliefs about ensoulment and the inception of personhood, and science can neither prove nor refute the teaching of those religions that consider the zygote to be a human person with an immortal soul. In his
Research into human development involves the use of human embryos and their derivative cells and tissues. How religions view the human embryo depends on beliefs about ensoulment and the inception of personhood, and science can neither prove nor refute the teaching of those religions that consider the zygote to be a human person with an immortal soul. In his  MicroRNAs (miRNAs) contribute to the progressive changes in gene expression that occur during development. In their
MicroRNAs (miRNAs) contribute to the progressive changes in gene expression that occur during development. In their 
 (1 votes)
(1 votes)