In Development this week (Vol. 141, Issue 10)
Posted by Seema Grewal, on 6 May 2014
Here are the highlights from the current issue of Development:
Sara sorts out stem cell asymmetric division
 Adult stem cells play crucial roles in tissue homeostasis, giving rise to both new stem cells and differentiating daughter cells. The generation of these two cell types often involves the asymmetric distribution of cell fate determinants, but how these factors are partitioned asymmetrically has been unclear. Now (p. 2014), Chrystelle Montagne and Marcos Gonzalez-Gaitan reveal a role for Sara endosomes in the asymmetric division of Drosophilaintestinal stem cells (ISCs). Using live imaging of the adult fly midgut, the researchers first show that Sara endosomes, which are characterised by the localisation of the endosomal protein Sara, are unequally partitioned during ISC divisions, being preferentially targeted to the presumptive differentiating cell. They further examine the distribution of Notch and Delta, which have been implicated in regulating ISC fate, and show that both Notch and Delta traffic through Sara endosomes and, accordingly, are also asymmetrically dispatched to the differentiating cell. Importantly, they demonstrate that midgut homeostasis is perturbed in Sara mutants; the number of ISCs in the midgut is significantly higher in Sara mutants than in controls, indicating that Sara endosomes play a central role in assigning ISC fate. These, together with other findings, uncover a cell-intrinsic endosomal-based mechanism for regulating cell fate and asymmetric cell division.
Adult stem cells play crucial roles in tissue homeostasis, giving rise to both new stem cells and differentiating daughter cells. The generation of these two cell types often involves the asymmetric distribution of cell fate determinants, but how these factors are partitioned asymmetrically has been unclear. Now (p. 2014), Chrystelle Montagne and Marcos Gonzalez-Gaitan reveal a role for Sara endosomes in the asymmetric division of Drosophilaintestinal stem cells (ISCs). Using live imaging of the adult fly midgut, the researchers first show that Sara endosomes, which are characterised by the localisation of the endosomal protein Sara, are unequally partitioned during ISC divisions, being preferentially targeted to the presumptive differentiating cell. They further examine the distribution of Notch and Delta, which have been implicated in regulating ISC fate, and show that both Notch and Delta traffic through Sara endosomes and, accordingly, are also asymmetrically dispatched to the differentiating cell. Importantly, they demonstrate that midgut homeostasis is perturbed in Sara mutants; the number of ISCs in the midgut is significantly higher in Sara mutants than in controls, indicating that Sara endosomes play a central role in assigning ISC fate. These, together with other findings, uncover a cell-intrinsic endosomal-based mechanism for regulating cell fate and asymmetric cell division.WNT takes a free ride
 It is widely accepted that, in amniotes, WNTs secreted by the dorsal neural tube form a concentration gradient that regulates somite patterning and myotome organisation. Here, Olivier Serralbo and Christophe Marcelle challenge this assumption and uncover a novel mode of long-range WNT signalling in which WNTs are delivered to their target sites by migratory neural crest cells (p. 2057). The researchers first show that WNT1 protein is present on the surface of early migrating neural crest cells (NCCs) in the chick embryo. Furthermore, they demonstrate that the migration of NCCs is required for correct myotome organisation and for the WNT1-dependent activation of WNT11 in a somite derivative known as the dorsomedial lip (DML). These processes, in turn, are dependent on expression of the heparin sulphate proteoglycan GPC4 by NCCs; knockdown of GPC4 in NCCs, but not in DML cells, causes a reduction in WNT11 expression in the DML, highlighting a crucial role for GPC4 in donor but not receiving cells. Overall, these findings suggest a model in which WNT proteins are loaded onto migratory NCCs and are physically delivered to the receiving cells of the DML in a GPC4-dependent manner.
It is widely accepted that, in amniotes, WNTs secreted by the dorsal neural tube form a concentration gradient that regulates somite patterning and myotome organisation. Here, Olivier Serralbo and Christophe Marcelle challenge this assumption and uncover a novel mode of long-range WNT signalling in which WNTs are delivered to their target sites by migratory neural crest cells (p. 2057). The researchers first show that WNT1 protein is present on the surface of early migrating neural crest cells (NCCs) in the chick embryo. Furthermore, they demonstrate that the migration of NCCs is required for correct myotome organisation and for the WNT1-dependent activation of WNT11 in a somite derivative known as the dorsomedial lip (DML). These processes, in turn, are dependent on expression of the heparin sulphate proteoglycan GPC4 by NCCs; knockdown of GPC4 in NCCs, but not in DML cells, causes a reduction in WNT11 expression in the DML, highlighting a crucial role for GPC4 in donor but not receiving cells. Overall, these findings suggest a model in which WNT proteins are loaded onto migratory NCCs and are physically delivered to the receiving cells of the DML in a GPC4-dependent manner.
ELAVating insight into Hox RNA processing
 Hox genes play a crucial role in assigning cellular identities along the anterior-posterior axis of animal bodies. Hox gene expression can be regulated via transcriptional mechanisms and recent studies have also uncovered a regulatory role for Hox RNA processing, yet the mechanisms underlying this regulation remain unknown. Now, Claudio Alonso and colleagues identify the neural RNA-binding protein ELAV as a key regulator of Hox RNA processing in the Drosophila embryonic central nervous system (p. 2046). The researchers use the Drosophila Hox gene Ultrabithorax (Ubx) as a gene model for investigating RNA processing and discover that elav mutants produce patterns of Ubx alternative splicing and polyadenylation that are distinct from those observed in wild-type embryos. They further demonstrate that ELAV binds directly to discrete elements within Ubx RNA. In the absence of elav, they report, Ubx mRNA and protein levels are reduced, whereas nascent Ubx RNA transcripts accumulate, suggesting that ELAV-dependent processing of Ubx RNA is able to fine-tune the levels of Ubx expressed. Furthermore, an analysis of the cellular pathways affected in elav mutants reveals a role for ELAV in Hox-dependent apoptosis. The authors thus propose a model in which ELAV modulates Hox RNA processing, expression and function in order to regulate local programmes of neural differentiation.
Hox genes play a crucial role in assigning cellular identities along the anterior-posterior axis of animal bodies. Hox gene expression can be regulated via transcriptional mechanisms and recent studies have also uncovered a regulatory role for Hox RNA processing, yet the mechanisms underlying this regulation remain unknown. Now, Claudio Alonso and colleagues identify the neural RNA-binding protein ELAV as a key regulator of Hox RNA processing in the Drosophila embryonic central nervous system (p. 2046). The researchers use the Drosophila Hox gene Ultrabithorax (Ubx) as a gene model for investigating RNA processing and discover that elav mutants produce patterns of Ubx alternative splicing and polyadenylation that are distinct from those observed in wild-type embryos. They further demonstrate that ELAV binds directly to discrete elements within Ubx RNA. In the absence of elav, they report, Ubx mRNA and protein levels are reduced, whereas nascent Ubx RNA transcripts accumulate, suggesting that ELAV-dependent processing of Ubx RNA is able to fine-tune the levels of Ubx expressed. Furthermore, an analysis of the cellular pathways affected in elav mutants reveals a role for ELAV in Hox-dependent apoptosis. The authors thus propose a model in which ELAV modulates Hox RNA processing, expression and function in order to regulate local programmes of neural differentiation.Ssm1b: a novel modifier of DNA methylation
 A locus in mice known as strain-specific modifier 1 (Ssm1) has previously been shown to be responsible for the strain-dependent methylation of E. coli gpt-containing transgenic sequences. Now, Ursula Storb and co-workers identify the Ssm1b gene that underlies this phenotype and characterise its expression in early mouse embryos (p. 2024). Through extensive mapping studies, the researchers identify Ssm1b as a KRAB-zinc finger gene that is located on distal chromosome 4. They further demonstrate that Ssm1b is expressed in early embryos up until embryonic day 8.5 and, in line with this, its target transgene gains partial methylation by this stage. The Ssm1b gene lacks the conserved transferase sequence present in all DNA methyltransferases, but the researchers demonstrate that Ssm1b mediates transgene methylation via the de novo methyltransferase Dnmt3b. By contrast, they report, the methylated DNA-binding protein Mecp2 is not involved in Ssm1b-dependent DNA methylation. These findings, together with preliminary analyses of Ssm1b function, uncover a novel gene and highlight the existence of a new family of genes that can initiate DNA methylation and chromatin modification and hence are likely to be involved in the epigenetic control of early development.
A locus in mice known as strain-specific modifier 1 (Ssm1) has previously been shown to be responsible for the strain-dependent methylation of E. coli gpt-containing transgenic sequences. Now, Ursula Storb and co-workers identify the Ssm1b gene that underlies this phenotype and characterise its expression in early mouse embryos (p. 2024). Through extensive mapping studies, the researchers identify Ssm1b as a KRAB-zinc finger gene that is located on distal chromosome 4. They further demonstrate that Ssm1b is expressed in early embryos up until embryonic day 8.5 and, in line with this, its target transgene gains partial methylation by this stage. The Ssm1b gene lacks the conserved transferase sequence present in all DNA methyltransferases, but the researchers demonstrate that Ssm1b mediates transgene methylation via the de novo methyltransferase Dnmt3b. By contrast, they report, the methylated DNA-binding protein Mecp2 is not involved in Ssm1b-dependent DNA methylation. These findings, together with preliminary analyses of Ssm1b function, uncover a novel gene and highlight the existence of a new family of genes that can initiate DNA methylation and chromatin modification and hence are likely to be involved in the epigenetic control of early development.
Plus…
Adult neurogenesis: mechanisms and functional significance
 Adult neurogenesis has been implicated in physiological brain function, and failing or altered neurogenesis has been associated with a number of neuropsychiatric diseases. Simon Braun and Sebastian Jessberger provide an overview of the mechanisms governing the neurogenic process in the adult brain and describe how new neurons may contribute to brain function in health and disease. See the Development at a Glance poster article on p. 1983
Adult neurogenesis has been implicated in physiological brain function, and failing or altered neurogenesis has been associated with a number of neuropsychiatric diseases. Simon Braun and Sebastian Jessberger provide an overview of the mechanisms governing the neurogenic process in the adult brain and describe how new neurons may contribute to brain function in health and disease. See the Development at a Glance poster article on p. 1983
Apical constriction: themes and variations on a cellular mechanism driving morphogenesis
 Apical constriction is a cell shape change that promotes tissue remodelling in a variety of contexts. Martin and Goldstein review the cellular machinery required for apical constriction and discuss how it can be tunedto regulate apical constriction in diverse cellular contexts. See the Review article on p. 1987
Apical constriction is a cell shape change that promotes tissue remodelling in a variety of contexts. Martin and Goldstein review the cellular machinery required for apical constriction and discuss how it can be tunedto regulate apical constriction in diverse cellular contexts. See the Review article on p. 1987
Cell migration: from tissue culture to embryos
 Cell migration is a fundamental process that occurs during embryo development. Here, Concha and colleagues review the guidance principles of in vitro cell locomotion and examine how they apply to examples of directed cell migration observed in vivo during development. See the Review on p. 1999
Cell migration is a fundamental process that occurs during embryo development. Here, Concha and colleagues review the guidance principles of in vitro cell locomotion and examine how they apply to examples of directed cell migration observed in vivo during development. See the Review on p. 1999


 (No Ratings Yet)
(No Ratings Yet)
 (1 votes)
(1 votes)





 Mesoangioblasts (MABs) are progenitor cells of embryonic derivation with mesodermal potential. They have been successfully used to restore skeletal muscle loss in dystrophic mice, but despite the clinical potential of these cells, their origin and role during development has not been defined. Now, on p.
Mesoangioblasts (MABs) are progenitor cells of embryonic derivation with mesodermal potential. They have been successfully used to restore skeletal muscle loss in dystrophic mice, but despite the clinical potential of these cells, their origin and role during development has not been defined. Now, on p.  The assembly of the peripheral nervous system occurs in a precise order: motor efferent axons (MEs) emerge first, followed by somatosensory afferent axons (SAs), and then by sympathetic efferent axons (SEs). While this order is clearly defined, it is not clear whether the pioneering axons provide instructive cues for the trailing axons to follow, and thus whether the network represents a true hierarchy. In this issue (p.
The assembly of the peripheral nervous system occurs in a precise order: motor efferent axons (MEs) emerge first, followed by somatosensory afferent axons (SAs), and then by sympathetic efferent axons (SEs). While this order is clearly defined, it is not clear whether the pioneering axons provide instructive cues for the trailing axons to follow, and thus whether the network represents a true hierarchy. In this issue (p.  How does a developing tissue know how much to grow and when to stop? On p.
How does a developing tissue know how much to grow and when to stop? On p.  Wound repair is a fundamental process that is required for tissue homeostasis and regeneration following damage. Most studies of wound healing have focussed on changes in the leading edge of wounded cells, but here William Razzell, Will Wood and Paul Martin show that morphogenetic cell shape changes that occur multiple cell rows back from the wound are important for efficient wound re-epithelialisation (p.
Wound repair is a fundamental process that is required for tissue homeostasis and regeneration following damage. Most studies of wound healing have focussed on changes in the leading edge of wounded cells, but here William Razzell, Will Wood and Paul Martin show that morphogenetic cell shape changes that occur multiple cell rows back from the wound are important for efficient wound re-epithelialisation (p.  Synaptogenesis is a complex process that involves the coordinated assembly of pre- and postsynaptic compartments. Various extracellular pathways and cues have been shown to regulate synapse formation but here, on p.
Synaptogenesis is a complex process that involves the coordinated assembly of pre- and postsynaptic compartments. Various extracellular pathways and cues have been shown to regulate synapse formation but here, on p.  Tissue morphogenesis is driven by coordinated cellular deformations and recent studies have shown that these changes in cell shape are powered by intracellular contractile networks comprising actin filaments, actin cross-linkers and myosin motors. In their Development at a Glance poster article, Munjal and Lecuit provide an overview of the mechanics, principles and regulation of actomyosin-driven cellular tension driving tissue morphogenesis. See the article on p.
Tissue morphogenesis is driven by coordinated cellular deformations and recent studies have shown that these changes in cell shape are powered by intracellular contractile networks comprising actin filaments, actin cross-linkers and myosin motors. In their Development at a Glance poster article, Munjal and Lecuit provide an overview of the mechanics, principles and regulation of actomyosin-driven cellular tension driving tissue morphogenesis. See the article on p. 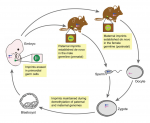 Genes that are subject to genomic imprinting in mammals are preferentially expressed from a single parental allele. These imprinted genes can directly regulate fetal growth, and recent work has also demonstrated intricate roles for imprinted genes in the brain and in induced pluripotent stem cells and adult stem cells. As Bartolomei and colleagues review here, these findings highlight the complex nature and developmental importance of imprinted genes. See the Review on p.
Genes that are subject to genomic imprinting in mammals are preferentially expressed from a single parental allele. These imprinted genes can directly regulate fetal growth, and recent work has also demonstrated intricate roles for imprinted genes in the brain and in induced pluripotent stem cells and adult stem cells. As Bartolomei and colleagues review here, these findings highlight the complex nature and developmental importance of imprinted genes. See the Review on p.