by Jessica Chen and Jenna Galloway
Animals can contort their bodies into a diversity of movements: running, jumping, climbing, and swimming to name a few. All of these movements are possible because tendons transmit the force produced by the muscles to the bones. Most of us do not pay much attention to our tendons and ligaments until something happens to them. Sports and repetitive motion injuries are very common yet are complicated by slow and limited repair. Surprisingly, very little is known about how tendons and ligaments form and organize to make the appropriate connections within the musculoskeletal system, and then, maintain and repair themselves in the adult. Part of our limited knowledge concerning their developmental program had been due to the absence of early markers at stages that preceded the morphological detection of tendons. The identification of the transcription factor Scleraxis (scx)as a robust marker of tendon and ligament progenitors provided the means to gain an understanding of the molecular regulators of tendon cell induction and organization (Schweitzer et al., 2001). With these questions in mind, we developed the zebrafish as a model to study tendon biology in our recent paper in Development.
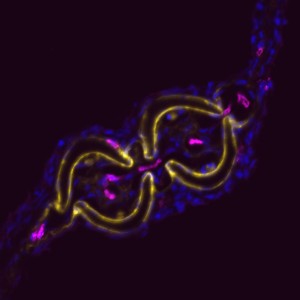
Why study zebrafish and do they have tendons? When we began these studies, we were often asked this question, and itforced us to consider if zebrafish would be an appropriate model to study tendon and ligament biology. Clearly, the forces fish experience in an aquatic habitat are much different than those felt by terrestrial land animals. As such, the tendons in these diverse species, which perform very different movements, could also be different. Previous molecular studies of zebrafish tendon tissues have focused on the myosepta, which connects the muscle segments along the body axis enabling undulatory swimming (Bassett et al., 2003; Charvet et al., 2013; Kudo et al., 2004). The myoseptal tissue functions as a tendon in transmitting force necessary for swimming, and we found that it expresses many tendon markers during developmental stages. At these stages, however, it primarily connects muscle to muscle, and in adult zebrafish, its structure is not similar to the linear tendons of mammals (Charvet et al., 2011; Summers and Koob, 2002). In contrast, we focused most of our studies on the cranial region when we began examining tendon markers in zebrafish embryos. We concentrated on this anatomical location for two principal reasons: here, cartilage and bone are primarily found developing in close proximity to muscle, and second, the pressure for prey capture and feeding would require a functioning musculoskeletal apparatus at very early stages. Indeed, it was in the cranial regions that we found the co-expression of many tendon markers, including scleraxisa, and also where, in adults, the tissue was similar on the ultrastructural level to that of the linear tendons of mammals.

In demonstrating that the zebrafish cranial tendon populations are homologous to their mammalian counterparts, we have expanded our ability to study this tissue in the context of musculoskeletal patterning. Although our work was the first to molecularly describe the head tendons and ligaments in zebrafish, the cranial musculoskeletal anatomy and functional morphology of ray-finned fishes has been studied for over a century (reviewed in (Ferry-Graham and Lauder, 2001)), and models of feeding mechanics in adult fish have detailed cranial tendon and ligament attachments in the context of their role during jaw movements (Liem, 1967; Westneat, 1990). During ontogeny, efficient feeding during developmental stages is linked to survival and thought to be evolutionarily advantageous (Houde and Schekter, 1980). We found it striking that the regions with robust co-expression of tendon markers coincided with pivotal points of force transmittance during larval feeding (Hernandez et al., 2002). Given the diversity of craniofacial morphology in teleosts, it would be interesting to understand how tendon and ligament progenitor induction and organization may play a role in shaping the cranial musculoskeletal anatomy. It is known that the neural crest-derived connective and skeletal tissues pattern the cranial muscle attachments in avian systems (Noden, 1986; Noden, 1988). Our work suggests that cartilage may have a role in tendon organization, and previous studies have found that a disruption to cartilage development results in distorted muscle shapes (Yan et al., 2002). Together, these results underscore the importance of tendon-cartilage interactions in musculoskeletal patterning, and future work in the zebrafish will begin to dissect the mechanisms underlying these processes.
Ultimately, we believe that the fish will provide new avenues for studying tendon and ligament biology in a powerful vertebrate genetic system. Findings from chemical screens in zebrafish have already demonstrated the potential for clinical translation in the treatment of a variety of human disease and developmental conditions (Bowman and Zon, 2010; Kaufman et al., 2009). Identifying the pathways that regulate tendon progenitor cell induction, growth, differentiation, and the formation of the attachment sites has relevance in the clinical setting where poor healing, scar tissue and high failure rates at the tendon-bone interface are quite common. The zebrafish system has the potential to not only expand our knowledge of the mechanisms underlying tendon formation and organization through the use of live-imaging and screen based approaches, but also the ability through the creation of injury and disease models and the development of drug discovery platforms to impact clinical therapies.
References
Bassett, D., Bryson-Richardson, R. J., Daggett, D. F., Gautier, P., Keenan, D. G., & Currie, P. D. (2003). Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo Development, 130 (23), 5851-5860 DOI: 10.1242/dev.00799
Bowman TV, & Zon LI (2010). Swimming into the future of drug discovery: in vivo chemical screens in zebrafish. ACS chemical biology, 5 (2), 159-61 PMID: 20166761
Charvet, B., Guiraud, A., Malbouyres, M., Zwolanek, D., Guillon, E., Bretaud, S., Monnot, C., Schulze, J., Bader, H., Allard, B., Koch, M., & Ruggiero, F. (2013). Knockdown of col22a1 gene in zebrafish induces a muscular dystrophy by disruption of the myotendinous junction Development, 140 (22), 4602-4613 DOI: 10.1242/dev.096024
Charvet, B., Malbouyres, M., Pagnon-Minot, A., Ruggiero, F., & Guellec, D. (2011). Development of the zebrafish myoseptum with emphasis on the myotendinous junction Cell and Tissue Research, 346 (3), 439-449 DOI: 10.1007/s00441-011-1266-7
Ferry-Graham, L., & Lauder, G. (2001). Aquatic prey capture in ray-finned fishes: A century of progress and new directions Journal of Morphology, 248 (2), 99-119 DOI: 10.1002/jmor.1023
Hernandez, L., Barresi, M. J., & Devoto, S. H. (2002). Functional Morphology and Developmental Biology of Zebrafish: Reciprocal Illumination from an Unlikely Couple Integrative and Comparative Biology, 42 (2), 222-231 DOI: 10.1093/icb/42.2.222
Houde, E., & Schekter, R. (1980). Feeding by marine fish larvae: developmental and functional responses Environmental Biology of Fishes, 5 (4), 315-334 DOI: 10.1007/BF00005186
Kaufman, C., White, R., & Zon, L. (2009). Chemical genetic screening in the zebrafish embryo Nature Protocols, 4 (10), 1422-1432 DOI: 10.1038/nprot.2009.144
Kudo, H., Amizuka, N., Araki, K., Inohaya, K., & Kudo, A. (2004). Zebrafish periostin is required for the adhesion of muscle fiber bundles to the myoseptum and for the differentiation of muscle fibers Developmental Biology, 267 (2), 473-487 DOI: 10.1016/j.ydbio.2003.12.007
Liem KF (1967). Functional morphology of the head of the anabantoid teleost fish Helostoma temmincki. Journal of morphology, 121 (2), 135-58 PMID: 6034528
Noden, D. (1986). Patterning of avian craniofacial muscles Developmental Biology, 116 (2), 347-356 DOI: 10.1016/0012-1606(86)90138-7
Noden DM (1988). Interactions and fates of avian craniofacial mesenchyme. Development (Cambridge, England), 103 Suppl, 121-40 PMID: 3074905
Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, & Tabin CJ (2001). Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development, 128 (19), 3855-66 PMID: 11585810
Summers, A., & Koob, T. (2002). The evolution of tendon — morphology and material properties Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 133 (4), 1159-1170 DOI: 10.1016/S1095-6433(02)00241-6
Westneat, M. (1990). Feeding mechanics of teleost fishes (Labridae; Perciformes): A test of four-bar linkage models Journal of Morphology, 205 (3), 269-295 DOI: 10.1002/jmor.1052050304
Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, Chung BC, Westerfield M, Haffter P, Hopkins N, Kimmel C, & Postlethwait JH (2002). A zebrafish sox9 gene required for cartilage morphogenesis. Development, 129 (21), 5065-5079 PMID: 12397114
 (1 votes)
(1 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)

 (3 votes)
(3 votes)