Meet the Node correspondents: Alexandra Bisia
Posted by Reinier Prosee, on 19 April 2023
We recently announced that we will be working with three newly appointed the Node correspondents, who will be helping us to develop and write content for the Node in 2023. We caught up with each of them to chat about their research backgrounds and the topics that they’re excited to write about over the course of the coming year.

Working alongside Brent Foster, we have Alexandra Bisia who is currently a DPhil student in the lab of Elizabeth Robertson at the Sir William Dunn School of Pathology, University of Oxford. In fact, you may have already seen her first post as a correspondent in which she covers the Cut + Paste exhibition currently on display in the Francis Crick Institute, London. Alex won the 2020 BSCB Science Writing Prize and here we discuss how she would like to channel her creative energy into the correspondent role. Also, we cover her (international) career path up till now and her love of languages (and word games!).
Congratulations on being selected as one of our new the Node correspondents. How did you hear about the role, and why did you decide to apply for it?
I’ve been following the Node for a while – in fact The Company of Biologists in general – and really like the science- and scientist-centric approach. What I like about the Node is the diversity of the contributors who all have something interesting thing to say. I also enjoy the Tsuku Mogami series of short stories published in Development, which use animated objects to illustrate biological concepts and findings. It’s so creative, and yet so scientific – really inspiring!
So when I saw that the Node was looking for correspondents, I honestly filled in the application within ten minutes or so. I didn’t want to overthink it – if it was meant to be, it was meant to be. I’m thrilled that this turned out to be the case.
Have you done much science writing before, or will this (mostly) be a new experience for you?
Well, not in a very formal way, or within a particular framework. I have written some pieces for my department website (the Sir William Dunn School of Pathology at the University of Oxford). These were either departmental news stories or an overview of papers that were recently published by people working in the department. One thing that perhaps stands out for me is the piece that I submitted for the writing competition of the British Society for Cell Biology. It was three years ago, at the start of my PhD and also at the start of the COVID-19 lockdown, when I had all this energy and very little to do with it. So I poured all that into writing and got really creative with it. I wrote about the evolution of multicellularity and had a lot of fun doing that. It’d be great to bring back some of that creative energy into the correspondent role.
What sort of topics are you excited to write about for the Node? Do they relate to your own work, or are you looking to branch out into other areas?
I’d say that this is a great opportunity to look at something outside of my direct area of research. The ideas that I’ve come up with so far include non-model organism and recent breakthroughs in the medical sciences. As a developmental biologist by training, this will be a unique chance to reach out to people that I normally wouldn’t get to meet.
Actually, I already got to do somethings outside of my normal routine while preparing my first correspondents’ post. I went to report on the Cut + Paste exhibition at the Francis Crick Institute. I’ve never given much thought to what goes into organising an exhibition and it was fun to get a glimpse behind the curtain. I talked to Dr Güneş Taylor, one of the exhibition’s scientific advisors, and heard from the creative consultancy that took on the design of the exhibition. I was the only person there with a notebook, just jotting stuff down. I felt like a real dork, but it was a really fun experience.
Could you tell us a bit more about your own research? What has your career path been like so far?
I was born in Athens and raised in Crete, which is where I went to school. The science teachers at my school, especially for biology and chemistry, were really engaging, which influenced my later choices. I decided to go to Edinburgh for my undergraduate degree, as I had learned all about Dolly the sheep. Besides my scientific education though, I learned that Scotland is just a one-of-a-kind, special place. It’s such a beautiful country to live in!

As part of my undergraduate, I also studied at UC Berkeley for a year, which was a lot of fun. In Edinburgh, though, there was one Professor – Prof. Jamie Davies – who during one of his lectures convinced me that I wanted to study developmental biology. I knew it within the span of 50 minutes, I was like “this is my life now”. I ended up doing my thesis project in his lab, followed by a DPhil (PhD) in Chromosome and Developmental Biology here in Oxford, in Liz Robertson’s lab.
What has it been like to live and work in so many different places?
It’s been absolutely lovely to get the opportunity to make new homes. Of course, it also means that your heart gets broken each time you have to leave again. But then you do always have this second, and third, and fourth home. It’s a really enriching experience and I already find myself thinking where I will go next, and how that will make me feel.
For now, what are you hoping to gain from the experience of being a the Node correspondent?
I’m hoping to meet people with similar interests and see how they approach their job. I’d like to learn from their experience and take some of this back into my own work – my scientific work, but also my writing. My aim is to develop a writing style that is informative and entertaining for as many people as possible. There is already enough literature out there that’s exclusively comprehensible by scientists.
I’d like to think that I’ll always be doing some writing – whether full-time or as a hobby. Ideally, I’ll be working in research for a while longer and being part of the Node will be a great way to make new connections with people I have a lot in common with and whose experience can guide my future steps.
Finally, what do you enjoy doing in your spare time?
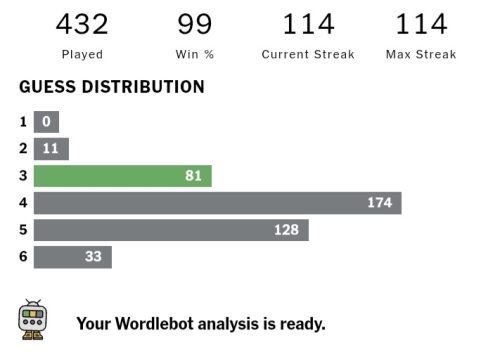
I love word games – I’m currently on a 114-day Wordle streak! I guess it comes from my love of languages. I speak Greek, English, French, Spanish, and ‘baby’ Arabic. I have Arabic-speaking family and I thought it was a good reason for me to learn the language and so – during my first year in Oxford – I would go up to the Center for Islamic Studies for a language class once a week. It was a lot of fun, but it turns out it that I now sound like a cross between a news presenter and a cartoon every time I speak it. It’s because the Arabic they teach you isn’t actually spoken by anyone – it’s the formal version which people don’t use in real life. Still, most people shower you with praise if you can even speak five words in their language, which is always a great feeling.
Note: You can find all the posts from the Node correspondents here: https://thenode.biologists.com/the-node-correspondents/


 (3 votes)
(3 votes)
 (No Ratings Yet)
(No Ratings Yet)