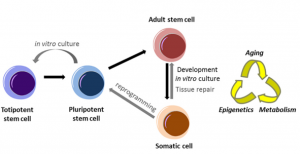
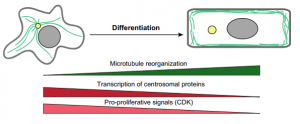
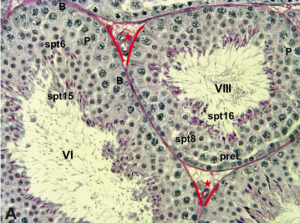
Developmental Biology enquires about the fundamental processes that underpin the fertilisation of an egg cell and its step-by-step transformation into the fascinating complexity of a whole organism (Box 1).
Box 1:
Some definitions of Developmental Biology
- Developmental Biology is the study of the processes by which organs grow and develop. Modern developmental biology studies the genetic control of cell growth, differentiation and morphogenesis, which is the process that gives rise to tissues, organs and anatomy, but also regeneration and ageing (after L. Wolpert)
- Developmental biology is the study of the process by which animals and plants grow and develop, and is synonymous with ontogeny (Wikipedia).
- Developmental Biology is the causal analysis of the cellular mechanisms that drive processes of growth, pattern formation and morphogenesis (A. Martínez Arias)
Further advocacy articles
- Development special advocacy articles
- Maartens, A., Prokop, A., Brown, K., Pourquié, O. (2018). Advocating developmental biology (Editorial). Development 145 — [LINK]
- Ebisuya, M., Briscoe, J. (2018). What does time mean in development? Development 145 — [LINK]
- Wiese, K. E., Nusse, R., van Amerongen, R. (2018). Wnt signalling: conquering complexity. Development 145 — [LINK]
- Philip W. Ingham. (2018). From Drosophila segmentation to human cancer therapy. Development 145 — [LINK]
- Prokop, A. (2018). What is Developmental Biology – and why is it important? Open Access Govern, in press — [LINK]
- Duronio, R. J., O’Farrell, P. H., Sluder, G., Su, T. T. (2017). Sophisticated lessons from simple organisms: appreciating the value of curiosity-driven research. Disease Models & Mechanisms 10, 1381-1389 — [LINK] – comment by A Martínez Arias [LINK2]
- Gilbert, S. F. (2017). Developmental biology, the stem cell of biological disciplines. PLoS Biology 15, e2003691 — [LINK] – comment by A Martínez Arias [LINK2]
- Zhang, F. (2017). A new age of discovery for biology. In “The World in 2018”. The Economist — [LINK]
- St Johnston, D. (2015). The renaissance of Developmental Biology. PLoS Biol 13, e1002149 — [LINK]
- Wassarman, P. M. (2016) Essays on Developmental Biology. Current Topics in Developmental Biology — [PartA] [PartB]
- Hines, P. J., Marx, J., Parks, S. (1994). Frontiers in development. Science 266, 561-564 — [LINK]
- Crick, F. (1977). Developmental biology. In “The encyclopedia of ignorance – life sciences” (R. Duncan, M. Weston-Smith, Eds.), Vol. 2, pp. 299-303. Pergamon Press, Oxford, New York, Toronto, Sydney, Paris, Frankfurt — [LINK]
At first sight, Developmental Biology could be viewed as an academic discipline driven by mere curiosity and, hence, to be of little relevance to the big challenges of population health or sustainability. On the contrary, Developmental Biology – along with Physiology[1] – is arguably the most important biological discipline we have. Here we will explain and substantiate this statement[2].
(1) Developmental defects in humans are very abundant (Box 2). By studying the underlying mechanisms and causes, Developmental Biology addresses the key challenge of population health. Sustaining food resources is another major global challenge, and Developmental Biology can provide key strategies to improving crop and plant cultivation (see Mathan et al., 2016, Development 143, 3283ff. — LINK; further arguments will follow – please help us by contributing your ideas!).
Box 2: Statements from the literature illustrating the abundance of developmental defects in humans
- The frequency at which all classes of developmental defects occur is thought to be … exceeding half of initial pregnancies.
- Major developmental defects … occur in approximately 3% of live births.
- In 1995, major developmental defects accounted for approximately 70% of neonatal deaths (occurring before 1 month of age) and 22% of the 6,500 infant deaths (before 15 months of age) in the US.
- Approximately 30% of admissions to pediatric hospitals are for health problems associated with such defects.
source: Scientific Frontiers in Developmental Toxicology and Risk Assessment, 2000, National Academic Press, Washington DC, pp.354; edited by the National Research Council (LINK)
(2) Developmental Biology (like Physiology) is asking fundamental questions at the level of whole organisms, organs or tissues (Box 3). Notably, this is the level at which diseases become manifest. For this reason, Developmental Biology has been, and continues to be, most effective in delivering explanations for diseases or medically relevant processes including infertility, neonatal death, birth defects (e.g. deformation, body growth abnormalities, developmental brain disorders, blindness, deafness), cancer, wound healing, tissue regeneration (regenerative medicine including stem cell biology), etc.
Box 3: Fundamental questions asked by developmental biologists – and how they translate into biomedical application
- What processes lead to fertilisation and the initiation of development? How can we overcome infertility and childlessness?
- How do single fertilised egg cells, or later on groups of progenitor cells, generate the enormous cellular diversity of an organism and its organs and tissues? How do stem/progenitor cells generate whole tissues or organs – for example in regeneration or tissue engineering, and how does wound healing work?
- How do cells, which originate from common ancestors and contain the same genetic information, adopt different fates? How do cells change their identities and behaviours – for example in cancer?
- How do tissues and their cells know when to stop growing? How can cells evade growth control – for example in tumour growth?
- How is the formation of different cells/tissues coordinated in space and time? What are the patho-mechanisms underlying birth disorders?
(3) By asking fundamental questions at the level of organisms, organs and tissues, Developmental Biology-related research is a generator of new ideas and concepts (Box 4). These concepts essentially underpin the modern biomedical sciences and include cell signalling, tissue and body patterning, growth regulation, cell migration or morphogenesis; they form the basis for contemporary research into stem cells, cancer, wound healing, regeneration or ageing.
(4) Developmental Biology is exciting and powerful because it reaches across the different levels of biological complexity and explanation; phenomena at the level of organisms, organs or tissues can ultimately be understood only by tracing them back to events at the level of genes and cells. Consequently, Developmental Biology embraces disciplines such as genetics, molecular biology, (stem) cell biology, biochemistry, biophysics as well as evolutionary biology.
 (5) Developmental Biology capitalises on the principle of evolutionary conservation of genes, mechanisms and concepts by making informed and strategic uses of suitable model organisms, down to experimentally and genetically amenable invertebrates. The use of invertebrate model organisms provides an efficient and powerful strategy to generate new ideas, concepts and understanding which can then be tested (and often validated) in higher organisms, eventually leading to medical applications in humans (LINK). This discovery pipeline has been a central driver for the enormous contributions that Developmental Biology has made and continues to make to the biomedical sciences.
(5) Developmental Biology capitalises on the principle of evolutionary conservation of genes, mechanisms and concepts by making informed and strategic uses of suitable model organisms, down to experimentally and genetically amenable invertebrates. The use of invertebrate model organisms provides an efficient and powerful strategy to generate new ideas, concepts and understanding which can then be tested (and often validated) in higher organisms, eventually leading to medical applications in humans (LINK). This discovery pipeline has been a central driver for the enormous contributions that Developmental Biology has made and continues to make to the biomedical sciences.
Box 4: A metaphor explaining how Developmental Biology works
Understanding a combustion engine requires investigating its single parts, such as the sparking plug, cylinder or crank shaft. However, for a developmental biologist it is not sufficient to find out how each of these parts works, but this new knowledge needs to be linked back into the mechanistic framework that constitutes the entire engine and explains its function. Linking detailed findings back to the bigger question of understanding the engine is an important validation and filter step that reveals whether the detailed findings are actually relevant and make deeper sense. Only through such systematic and holistic “reverse enginieering” can the necessary systemic and conceptual understanding be achieved which is required to diagnose and eventually repair a faulty engine.
By executing all research work with the ultimate aim of understanding the bigger question (e.g. how to make an organ, appendix or entire body), Developmental Biology has become such an important contributor of concepts and understanding in the field of the biomedical and medical sciences.
As an interesting side note: already in the late 18th century, Johann Wolfgang von Goethe understood that it is not enough to “have the pieces in your hand” but that they need to be connected and “woven” together to “recognise and describe the living”.

These are only some thoughts (still incomplete and in need of further optimisation) about the importance of Developmental Biology as a discipline. If we want to improve the standing of our discipline, we MUST embrace these ideas with passion, help to complement and further improve them and, most importantly, use them whenever possible and adequate to advocate our discipline with self-confidence and enthusiasm. To do this with impact, we need to acknowledge that communicating science is a difficult task which requires belief, love for our subject, stamina, and efficient strategies that enable us to engage with a wide spectrum of target audiences. Such target audiences include the general public and schools, fellow scientists and clinicians, as well as politicians and other decision makers. But we need to be realistic and accept that most members of these target audiences will, by default, show little interest to engage with us. Therefore, intelligent and strategic long-term approaches – ideally shared within networks of scientists – likely provide the most promising path of engagement – as is explained in greater detail in the editorial of a special issue on science communication (published in Sem Cell Dev Biol; Box 5). For those who take an interest, this special issue provides many ideas of how long-term strategies for the communication of biomedical research can be implemented successfully – and can then even become rewarding for our own science and career. Please, also have a look at our link list of public engagement outreach resources which will provide you with further ideas and useful support materials.
Box 5:

Science communication in the field of fundamental biomedical research
(Vol. 70; Oct. 2017, free access for one year)
edited by Sam Illingworth & Andreas Prokop
The aim of this special issue is to provide concise and accessible advice on how to engage effectively, as well as share valuable practice gained from successful long-term science communication initiatives – thus providing ideas to all those who want to participate in dialogue with the public, policy makers or other scientists, or are participating already. This issue is primarily written for scientists working in the field of the biomedical sciences (and beyond), but will hopefully also be seen as a helpful resource for academic & professional science communicators. To appeal to both groups and hopefully stimulate impact-enhancing interdisciplinary collaborations, the issue is an unprecedented experiment written at the interface of both disciplines. The enormous opportunities of long-term approaches and the formation of interdisciplinary science communication networks are explained in great detail in our editorial. Further explanations about this issue were given in the recent NCCPE and PLoS|BLOGS posts.
———————————–

Cover art by Matt Girling. See figure legend in the editorial.
———————————–
1. EDITORIAL: Science communication in the field of fundamental biomedical research (Sam Illingworth, Andreas Prokop) – This article provides a critical assessment of the current state of science communication in the field of fundamental biomedical science, including lists of existing science communication initiatives, as well as a conceptual overview of the articles in the special issue [PDF] [see also].

2. Delivering effective science communication: advice from a professional science communicator (Sam Illingworth) – From his experience as Senior lecturer in Science communication, Sam Illingworth writes about important considerations for all who want to participate in active science communication. His tips and tricks for science fairs, school visits or many other forms of dialogue are provided in plain language. [PDF] [YouTube]

3. The nuts and bolts of evaluating science outreach (Suzanne Spicer) – From her experience as specialist for science communication at the Manchester Museum, Suzanne Spicer writes about the importance and practical implementation of project/event evaluation as an important means to improve quality and demonstrate impact. [PDF]

4. EuroStemCell: A European infrastructure for communication and engagement with stem cell research (Jan Barfoot, Kate Doherty, C. Clare Blackburn) – EuroStemCell is the flagship of non-commercial science communication in the area of the biomedical sciences, centred around their website with over a million annual visitors. The initiative was born out of an EU grant consortium and has developed into an interdisciplinary network that promotes responsible dialogue about stem cells, also through developing outreach resource and providing training. [PDF]

5. The Manchester Fly Facility: Implementing an objective-driven long-term science communication initiative (Sanjai Patel, Andreas Prokop) – The Manchester Fly Facility is an excellent example of an initiative communicating the importance of model organism biology. They promote Drosophila research through improved research training, school projects, science fair programs, and the development of publicly shared resources and strategies. Furthermore, they address Drosophila researchers to stimulate wider participation in science communication. [PDF]
6. Building dialogues between clinical and biomedical research through cross-species collaborations (Hsiao-Tuan Chao, Lucy Liu, Hugo J. Bellen) – Clinicians and biologists do not necessarily share a common scientific culture and way of thinking, but they share the common scientific goal of understanding and treating diseases. To this end, they can complement each other in powerful ways. This article by the Bellen lab provides helpful tools and successful strategies towards interdisciplinary collaboration between biologists working on model organisms and clinicians. [PDF]

7. DrosAfrica: Establishing a Drosophila community in Africa (Maria Dolores Martín-Bermudo, Luka Gebel, Isabel M. Palacios, I. M.) – DrosAfrica is an excellent example of how cost-effective, yet cutting edge research – as it is possible with Drosophila – can be used to free resources and promote active science and science education in disadvantaged countries. This article describes the origins of the DrosAfrica initiative and provides detailed insights into how their activities are organised. [PDF] [YouTube]

8. Engaging with primary schools: supporting the delivery of the new curriculum in evolution and inheritance (Paula Kover, Emily Hogge) – The “Teaching Evolution for Primary Children” initiative is an excellent example of successful dialogue with schools. The key strategy explained here is to research curricular and teacher requirements first, to then decide on suitable contents and develop useful resources in close collaboration with schools. This ensures influence of scientists on the correctness and quality of content and the influence of teachers on the way of presentation – in sum an excellent way to engage audiences at young age with science. [PDF]

9. The droso4schools project: long-term scientist-teacher collaborations to promote science communication and education in schools (Sanjai Patel, Sophie DeMaine, Joshua Heafield, Lynne Bianchi, Andreas Prokop) – Analogous to the previous article, the droso4schools initiative by the Manchester Fly Facility seeks true dialogue with teachers in that students are placed as teaching assistants in schools to learn about teaching strategies and teacher requirements. These experiences are then turned into sample lessons that use Drosophila as a teaching tool to explain curriculum-relevant concepts of biology, spiced up with micro experiments that are possible with flies even in classrooms. [PDF]

10. Science Communication at Scientific Societies (Jeanne Braha) – As former member of the AAAS science communication team, Jeanne Braha explains how societies can help their communities to improve science communication through offering advice, providing resources and training, connecting scientists to audiences, making information available to the public, and awarding prizes for outstanding science communication work. [PDF]
11. The Node and beyond – using social media in cell and developmental biology (Catarina Vicente, Aidan Maartens, Katherine Brown) – This article describes The Node as a unique communication platform for biomedical researchers, in particular from the fields of Cell and Developmental Biology. The Node does not only collate event and advocacy information, publish job vacancies and report about science-relevant topics, but it is also a blog site with international outreach where members of the science community can share news, thoughts or experiences. As such it acts as a modern electronic newsletter. [PDF]
[1] In the sense used here, Physiology comprises disciplines like immunology or functional studies in the field of neurobiology
[2] Arguments and examples given so far concern studies of animal development, and those for plant development will follow soon


 (No Ratings Yet)
(No Ratings Yet)
 (2 votes)
(2 votes)







































 (10 votes)
(10 votes) On p.
On p.  On p.
On p.  On p.
On p.