This editorial was first published in Development. We encourage feedback from the community on our policies – please leave any comments below.
A central premise of scientific publishing – that publication in a peer-reviewed journal means that the reader can be confident that an article is solid – has been challenged on a number of fronts in recent times. In October 2013, John Bohannon managed to get completely fictitious and nonsensical papers accepted for publication in over 100 Open Access journals that supposedly operated a peer-review system(Bohannon, 2013).Around the same time, an article in The Economist (http://www.economist.com/news/briefing/21588057-scientists-think-science-self-correcting-alarming-degree-it-not-trouble) suggested that biotech companies no longer trust the results of published studies from academia, finding them more often than not to be irreproducible (see also Begley and Ellis, 2012). And our own field has recently seen a number of high-profile retractions that have generated significant discussion about research oversight and the reliability of the publishing system for detecting research misconduct, as well as how such cases are handled by institutes, and the mainstream and social media.
Here at Development, and The Company of Biologists more broadly, we take our responsibilities to protect the integrity of the scientific record very seriously. We are members of the Committee on Publication Ethics (COPE) and follow their guidelines and policies in ethical matters (see www.publicationethics.org, for details). Like many other journals, we have a number of checks in place to try and detect potential ethical problems at the earliest possible stage – before publication. Unfortunately, no process is perfect (though we always aim to learn from any mistakes), and there are cases where published papers need to be investigated – and potentially corrected or retracted. Here again, we have clear procedures to ensure this is carried out as carefully, thoroughly and efficiently as possible (while maintaining confidentiality as appropriate), and we are constantly seeking to improve these procedures as policies change and new tools become available. Most of the ethical issues we encounter can be divided into three main categories: authorship disputes, plagiarism and inappropriate data manipulation.
Questions of authorship – who qualifies as an author, and in what order – are ideally agreed between authors well before submission, and adjusted if need be during the revision process. However, different individuals have different ideas as to what justifies authorship on a paper, and the increasingly collaborative nature of science means that assigning authorship is not always straightforward. Development’s guidelines state: ‘An author is someone who has made significant and substantial contributions to a study. This should include conception, design, execution and interpretation of the findings being published, and drafting and revising the article.’ Obviously, not all authors will necessarily have been involved with all stages of the work: consider the student who arrived halfway through a project – and so played no part in ‘conception’, or the lab head who didn’t actually carry out any of the bench work – the ‘execution’. We believe that a detailed ‘Author Contributions’ statement (required in all papers since 2013) is the best way of making it clear who did what on a paper, and in most cases, this works well. We also ensure that all listed authors are kept informed of the progress of a manuscript through our system, so that they can inform us of any potential concerns they may have.
But what about individuals, not listed on the paper, who believe they qualify for authorship? Such cases may only come to light after an article has been published, when the sometimes angry non-author emails us to assert their right to authorship. We aren’t in a position to weigh up the relative contributions of different individuals, so if the relevant parties can’t agree between themselves, we have to refer the case to the relevant institute(s) for them to investigate. This can be lengthy and painful, and serious authorship disputes can even result in papers being retracted from the literature. Our advice? Discuss authorship at an early stage in the project, be prepared to be flexible if contributions change (e.g. during revision, where someone new may have to step in to complete experiments – particularly if original authors have left the lab), keep lines of communication with collaborators and former colleagues open, and ensure that those who don’t quite qualify for authorship are recognised in the Acknowledgements section.
Fortunately, we have not experienced many problems with plagiarism in Development, although it is something we take seriously and police actively. All papers accepted for publication in the journal are run through a plagiarism detection program, iThenticate, that checks for significant matches to other papers or online material. We apply common sense here: there are only so many ways to describe a particular protocol, and original phrasing can be difficult particularly if English is not your native language. What we are looking for are cases where authors have clearly copied from another source without reference, and the degree of plagiarism defines our subsequent action: asking the authors to quote the original article, to rephrase their text, or – in extreme cases (which fortunately we have not yet encountered) – withdrawing the paper from publication. Of course, no software can detect so-called intellectual plagiarism, the ‘stealing’ of ideas, and here we rely on our reviewers and readers to alert us to potential problems, which we can then investigate.
By far the majority of ethical concerns we encounter involve data presentation and manipulation. The Journal of Cell Biology pioneered efforts to educate authors on appropriate figure processing (Rossner and Yamada, 2004) and to screen papers for possible problems before publication, and many publishers, including The Company of Biologists, now employ routine screening procedures to look for potentially inappropriate image manipulation in all accepted manuscripts (our policies on figure preparation can be found at http://dev.biologists.org/site/submissions/figure_prep.xhtml#manipulation). Referees can also play an important part here, by looking at figures with a critical eye when reviewing a paper and by alerting the editor to any potential concerns with data. Of course, it should be noted that an author who really wants to ‘cheat the system’ may be able to do so by clever use of Photoshop, or by manipulating the experiments conducted rather than the data collected. Moreover, our checks are not perfect, although we are always striving to improve them. However, when we do detect inconsistencies of concern – most frequently, unmarked splicing of gel lanes or alterations to the background of an image – we contact the authors, asking them to explain how the data were processed and to send us the original data behind the figure. In the vast majority of cases, authors are able to provide these easily and can reassure us that experiments have been conducted and reported appropriately, alterations can be made to the figure(s) where necessary, and there is no significant delay to publication. More substantial errors, such as duplicating data between panels or figures, can also be detected in some cases; it should be noted that these may be the result of honest error on the part of the author (the careless pasting in of the wrong image when preparing a complex panel) and can also be resolved rapidly. Unfortunately, however, not all instances of data manipulation are ‘innocent’, and we will not publish a paper where questions are hanging over the integrity of the data.
These are the cases that the reader never sees: those that are identified and resolved before publication. The more high-profile cases are those picked up after the paper has been published. Journals are receiving an increasing number of anonymous emails, often relating to papers published many years ago. These can be somewhat obtuse; a frequent complaint is that ‘error bars look too small or too similar to be real’. As responsible publishers, it is our duty to investigate all such complaints, and some real and important cases of image manipulation have been unearthed from anonymous tip-offs. We also receive reports from non-anonymous readers, as well as from the authors of the papers themselves – who may discover problems with their data as they follow it up in subsequent studies. The categories of errors and their frequencies are similar to those we identify pre-publication, as are our steps to investigate them. The first step is always to contact the corresponding author for an explanation; most issues can be readily resolved by an explanation from the authors and the provision of original data – potentially resulting in the publication of a Correction – but occasionally we do find more serious problems that may indicate intent to deceive and that require in-depth investigation.
So what do we do when we do identify serious problems – either before or after publication? As with authorship disputes, it is often impossible for us to resolve questions of data integrity at a distance. In these cases, we ask the appropriate bodies at the authors’ institute(s) to step in. They can look at the history of the research in detail, including going through lab notebooks, freezers and so on. This can take considerable time, although we will endeavour to keep our readers informed where appropriate: we are introducing a policy of publishing a Publisher’s Note to make readers aware of potential problems while investigations are ongoing. In all cases, we will take the necessary action once an investigation is complete to ensure the integrity of the scientific record. This might mean publishing a correction or a retraction.We are fortunate that retractions are rare here at Development, but this does not mean that we are reluctant to retract a paper where appropriate.
One important point to consider is the degree to which publishing policies have changed in the past decade or so. Those ‘manipulations’ that we now deem inappropriate (such as unmarked splicing of gel lanes) were common practice 10 years ago, and it can be unfair to judge the integrity of a paper published several years back by today’s standards. Moreover, manipulated data does not always imply fraudulent activity – authors frequently process their data for clarity or aesthetics without realising that this may not comply with journal policy. Still, many problems can be avoided through good recordkeeping and well-organised long-term storage of the original data (like many institutes, we expect that authors should retain records for a period of around 10 years), and through conservative post-processing of data – so that the submitted image accurately reflects the data gathered.
Simple mistakes can have significant consequences on the conclusions of a line of research. The vast majority of scientists are honest, and should be treated as such unless there is clear evidence to the contrary. Even in cases of clear misconduct, individuals should not be vilified – as an organisation, we take an educational rather than a punitive approach, and it is always important to retain perspective in these cases. It is often argued that the pressure to publish can lead researchers to cut corners, produce sloppy data or even commit fraud. This is no excuse, but in a culture where the rewards for a high-profile publication are so high, it is perhaps inevitable that a small number of people will succumb to these temptations. Fortunately, this is very rare and, at Development, we are proud to have the reputation of publishing papers that ‘stand the test of time’; for this, we are grateful to our editors, reviewers, authors and readers, whose careful work protects the integrity of our papers. Although there is always room for improvement, we hope that the policies we have in place, and continue to review and develop, will help to ensure that we correct any honest mistakes made and remain vigilant to the rare cases of intentional fraud.
Olivier Pourquié, Katherine Brown and Claire Moulton
References
Begley, C. G. and Ellis, L. M. (2012). Drug development: raise standards for preclinical cancer research. Nature 483, 531-533.
Bohannon, J. (2013). Who’s afraid of peer review? Science 342, 60-65.
Rossner, M. and Yamada, K. M. (2004). What’s in a picture? The temptation of image manipulation. J. Cell Biol. 166, 11-15.
 (1 votes)
(1 votes)
 Loading...
Loading...


 (3 votes)
(3 votes)
 (No Ratings Yet)
(No Ratings Yet)
 The mutated in molorectal cancer (Mcc) gene has been described as a tumour suppressor, and has been shown to interact with β-catenin and thus limit Wnt signalling. However, various data also indicate a potential role in regulating the cytoskeleton. Ray Dunn and colleagues set out to investigate this further in zebrafish and Xenopus (p.
The mutated in molorectal cancer (Mcc) gene has been described as a tumour suppressor, and has been shown to interact with β-catenin and thus limit Wnt signalling. However, various data also indicate a potential role in regulating the cytoskeleton. Ray Dunn and colleagues set out to investigate this further in zebrafish and Xenopus (p.  On p.
On p.  Meanwhile, Paul Knoepfler and co-workers investigate the role of the histone H3 variant H3.3 in regulating spermatogenesis (p.
Meanwhile, Paul Knoepfler and co-workers investigate the role of the histone H3 variant H3.3 in regulating spermatogenesis (p.  While most plants derive nitrogen (N) from the soil, some plants, such as legumes, can create their own supply by forming specialized root nodules that house N2-fixing bacteria. Nodule tissue ultimately derives from root cells that are reprogrammed to a nodule cell fate during nodule initiation, but exactly which root cells can contribute to the developing nodule primordial has not been fully established. Now, on p.
While most plants derive nitrogen (N) from the soil, some plants, such as legumes, can create their own supply by forming specialized root nodules that house N2-fixing bacteria. Nodule tissue ultimately derives from root cells that are reprogrammed to a nodule cell fate during nodule initiation, but exactly which root cells can contribute to the developing nodule primordial has not been fully established. Now, on p.  In May 2014, approximately 200 stem cell scientists from all over world gathered near Copenhagen in Denmark to participate in ‘The Stem Cell Niche’, part of the Copenhagen Bioscience Conferences series. The meeting covered an array of different stem cell systems from pluripotent stem cells and germ cells to adult stem cells of the lung, liver, muscle, bone and many more. In addition to the stem cell niche, the meeting focused on a number of cutting edge topics such as cell fate transitions and lineage reprogramming, as well as stem cells in ageing and disease, including cancer. Here, Kateri Moore and Giulio Cossu describe the exciting work that was presented and some of the themes that emerged from this excellent meeting. See the Meeting Review on p.
In May 2014, approximately 200 stem cell scientists from all over world gathered near Copenhagen in Denmark to participate in ‘The Stem Cell Niche’, part of the Copenhagen Bioscience Conferences series. The meeting covered an array of different stem cell systems from pluripotent stem cells and germ cells to adult stem cells of the lung, liver, muscle, bone and many more. In addition to the stem cell niche, the meeting focused on a number of cutting edge topics such as cell fate transitions and lineage reprogramming, as well as stem cells in ageing and disease, including cancer. Here, Kateri Moore and Giulio Cossu describe the exciting work that was presented and some of the themes that emerged from this excellent meeting. See the Meeting Review on p. 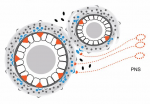 The hedgehog (HH) pathway is well known for its mitogenic and morphogenic functions during development, and HH signaling continues in discrete populations of cells within many adult mammalian tissues. Here, Ralitsa Petrova and Alex Joyner
The hedgehog (HH) pathway is well known for its mitogenic and morphogenic functions during development, and HH signaling continues in discrete populations of cells within many adult mammalian tissues. Here, Ralitsa Petrova and Alex Joyner 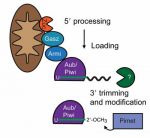 Since their discovery less than a decade ago, Piwi-interacting RNAs (piRNAs) have been shown to repress transposable elements in the germline and, hence, have been at the forefront of research aimed at understanding the mechanisms that maintain germline integrity. More recently, roles for piRNAs in gene regulation have emerged. In their Review, Eva-Maria Weick and Eric Miska highlight recent advances made in understanding piRNA function, highlighting the divergent nature of piRNA biogenesis in different organisms, and discussing the mechanisms of piRNA action during transcriptional regulation and in transgenerational epigenetic inheritance. See the Review on p.
Since their discovery less than a decade ago, Piwi-interacting RNAs (piRNAs) have been shown to repress transposable elements in the germline and, hence, have been at the forefront of research aimed at understanding the mechanisms that maintain germline integrity. More recently, roles for piRNAs in gene regulation have emerged. In their Review, Eva-Maria Weick and Eric Miska highlight recent advances made in understanding piRNA function, highlighting the divergent nature of piRNA biogenesis in different organisms, and discussing the mechanisms of piRNA action during transcriptional regulation and in transgenerational epigenetic inheritance. See the Review on p.