Here are the research highlights from the current issue of Development:
Evolution of mesoderm induction
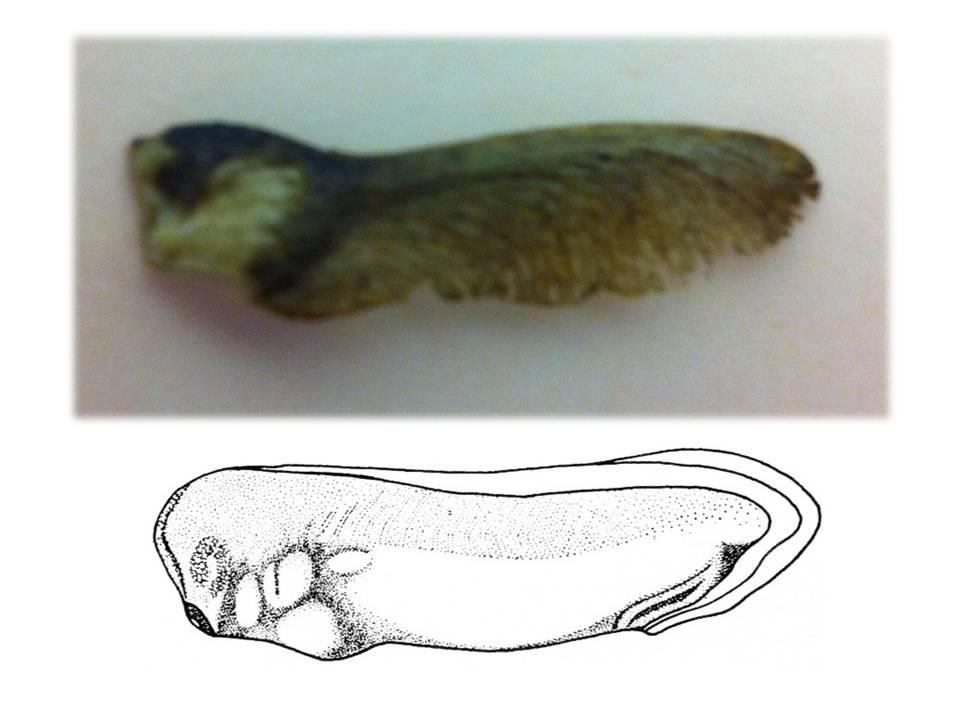
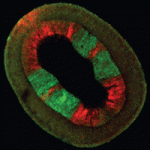 Fibroblast growth factors (FGFs) are essential for mesoderm induction in vertebrates and for early mesoderm formation in invertebrate chordates. However, functional studies to date do not support a role for FGF signalling in mesoderm induction in other deuterostomes (animals in which the first embryonic opening forms the anus), such as sea urchins. Thus, the ancestral role of FGF signalling during mesoderm specification in deuterostomes is unclear. On p. 1024, Christopher Lowe and co-workers examine the role of FGF signalling during the early development of the hemichordate Saccoglossus kowalevskii. The researchers report that the FGF ligand fgf8/17/18 is expressed in the ectoderm overlying sites of mesoderm specification within this marine worm’s archenteron (primitive gut) endomesoderm. Mesoderm induction, they show, requires contact between the ectoderm and the endomesoderm. Moreover, loss-of-function experiments indicate that FGF ligand and receptor are both necessary for mesoderm specification. These and other results indicate that FGF signalling is required throughout mesoderm specification in hemichordates and support an ancestral role for FGF signalling in mesoderm formation in deuterostomes.
Fibroblast growth factors (FGFs) are essential for mesoderm induction in vertebrates and for early mesoderm formation in invertebrate chordates. However, functional studies to date do not support a role for FGF signalling in mesoderm induction in other deuterostomes (animals in which the first embryonic opening forms the anus), such as sea urchins. Thus, the ancestral role of FGF signalling during mesoderm specification in deuterostomes is unclear. On p. 1024, Christopher Lowe and co-workers examine the role of FGF signalling during the early development of the hemichordate Saccoglossus kowalevskii. The researchers report that the FGF ligand fgf8/17/18 is expressed in the ectoderm overlying sites of mesoderm specification within this marine worm’s archenteron (primitive gut) endomesoderm. Mesoderm induction, they show, requires contact between the ectoderm and the endomesoderm. Moreover, loss-of-function experiments indicate that FGF ligand and receptor are both necessary for mesoderm specification. These and other results indicate that FGF signalling is required throughout mesoderm specification in hemichordates and support an ancestral role for FGF signalling in mesoderm formation in deuterostomes.
Histone demethylase builds testis niche
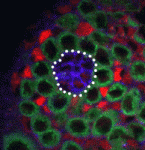 In adult stem cell niches, crosstalk between extrinsic cues (such as signals from neighbouring cells) and intrinsic cues (such as chromatin structure) regulates stem cell identity and activity. Now, on p. 1014, Xin Chen and colleagues report that the histone demethylase dUTX regulates crosstalk among the germline stem cells (GSCs), hub cells and cyst stem cells (CySCs) of the Drosophila testis niche. The researchers show that dUTX acts in CySCs to maintain hub cell identity by activating transcription of the Socs36E gene (which encodes an inhibitor of the JAK-STAT signalling pathway that is required for GSC identity and activity) via removal of a repressive histone modification near its transcription start site. dUTX also acts in GSCs, they report, to maintain hub structure through regulation of DE-cadherin, the Drosophila homologue of vertebrate cadherins. These results show how an epigenetic factor regulates crosstalk among different cell types within an adult stem cell niche and provide important information about the in vivo function of a histone demethylase.
In adult stem cell niches, crosstalk between extrinsic cues (such as signals from neighbouring cells) and intrinsic cues (such as chromatin structure) regulates stem cell identity and activity. Now, on p. 1014, Xin Chen and colleagues report that the histone demethylase dUTX regulates crosstalk among the germline stem cells (GSCs), hub cells and cyst stem cells (CySCs) of the Drosophila testis niche. The researchers show that dUTX acts in CySCs to maintain hub cell identity by activating transcription of the Socs36E gene (which encodes an inhibitor of the JAK-STAT signalling pathway that is required for GSC identity and activity) via removal of a repressive histone modification near its transcription start site. dUTX also acts in GSCs, they report, to maintain hub structure through regulation of DE-cadherin, the Drosophila homologue of vertebrate cadherins. These results show how an epigenetic factor regulates crosstalk among different cell types within an adult stem cell niche and provide important information about the in vivo function of a histone demethylase.
Novel Wnt inhibitors identified
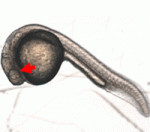 Members of the Eaf gene family are involved in tumour suppression and in embryogenesis but what are the molecular mechanisms that underlie these activities? Here (p. 1067), Wuhan Xiao and colleagues report that eaf1 and eaf2 modulate mesodermal and neural patterning in zebrafish embryos through inhibition of canonical Wnt/β-catenin signalling. They show that ectopic expression of eaf1 and eaf2 in zebrafish embryos and in cultured cells blocks β-catenin reporter activity. Furthermore, they show that Eaf1 and Eaf2 bind to the Armadillo repeat region and C-terminus of β-catenin, and to other β-catenin transcription complex proteins. Both the N- and C-terminus of Eaf1 and Eaf2 must be intact for their suppressive activity, they report. Finally, they show that the biological activities of Eaf family proteins are conserved across species. Together, these results identify a novel role for Eaf1 and Eaf2 in the inhibition of canonical Wnt/β-catenin signalling that might provide the mechanistic basis for the tumour suppressor activity of Eaf family proteins.
Members of the Eaf gene family are involved in tumour suppression and in embryogenesis but what are the molecular mechanisms that underlie these activities? Here (p. 1067), Wuhan Xiao and colleagues report that eaf1 and eaf2 modulate mesodermal and neural patterning in zebrafish embryos through inhibition of canonical Wnt/β-catenin signalling. They show that ectopic expression of eaf1 and eaf2 in zebrafish embryos and in cultured cells blocks β-catenin reporter activity. Furthermore, they show that Eaf1 and Eaf2 bind to the Armadillo repeat region and C-terminus of β-catenin, and to other β-catenin transcription complex proteins. Both the N- and C-terminus of Eaf1 and Eaf2 must be intact for their suppressive activity, they report. Finally, they show that the biological activities of Eaf family proteins are conserved across species. Together, these results identify a novel role for Eaf1 and Eaf2 in the inhibition of canonical Wnt/β-catenin signalling that might provide the mechanistic basis for the tumour suppressor activity of Eaf family proteins.
Linking planar polarity to junctional remodelling
 During morphogenesis, the elongation of polarised tissues involves cells within epithelial sheets and tubes making and breaking intercellular contacts in an oriented manner. How cells remodel their junctional contacts is poorly understood but growing evidence suggests that localised endocytic trafficking of E-cadherin might modulate cell adhesion. Now, Samantha Warrington and co-workers (p. 1045) report that the Frizzled-dependent core planar polarity pathway, which has been implicated in the regulation of cell adhesion through E-cadherin trafficking, promotes polarised cell rearrangements in Drosophila. The researchers report that the core planar polarity pathway promotes cell intercalation during tracheal tube morphogenesis by promoting E-cadherin turnover at junctions through local recruitment and regulation of the guanine exchange factor RhoGEF2. Core planar polarity pathway activity also leads to planar-polarised recruitment of RhoGEF2 and E-cadherin in the epidermis of the embryonic germband and the pupal wing. Thus, the researchers suggest, local promotion of E-cadherin endocytosis through recruitment of RhoGEF2 is a general mechanism by which the core planar polarity pathway promotes polarised cell rearrangements.
During morphogenesis, the elongation of polarised tissues involves cells within epithelial sheets and tubes making and breaking intercellular contacts in an oriented manner. How cells remodel their junctional contacts is poorly understood but growing evidence suggests that localised endocytic trafficking of E-cadherin might modulate cell adhesion. Now, Samantha Warrington and co-workers (p. 1045) report that the Frizzled-dependent core planar polarity pathway, which has been implicated in the regulation of cell adhesion through E-cadherin trafficking, promotes polarised cell rearrangements in Drosophila. The researchers report that the core planar polarity pathway promotes cell intercalation during tracheal tube morphogenesis by promoting E-cadherin turnover at junctions through local recruitment and regulation of the guanine exchange factor RhoGEF2. Core planar polarity pathway activity also leads to planar-polarised recruitment of RhoGEF2 and E-cadherin in the epidermis of the embryonic germband and the pupal wing. Thus, the researchers suggest, local promotion of E-cadherin endocytosis through recruitment of RhoGEF2 is a general mechanism by which the core planar polarity pathway promotes polarised cell rearrangements.
Signals for melanocyte stem cells
Adult stem cells are crucial for the growth, homeostasis and regeneration of adult tissues. Melanocyte (melanophore) stem cells (MSCs), which give rise to pigment cells in vertebrates, are an attractive model for studying the regulation of adult stem cells. In this issue, two papers provide new information about the involvement of signalling by the receptor tyrosine kinases Kit and ErbB in the establishment of MSCs in zebrafish.
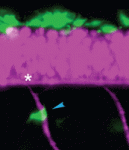 On p. 1003, Christiane Nüsslein-Volhard and colleagues investigate the embryonic origin of the melanophores that emerge during juvenile development and that contribute to the striking colour patterns of adult zebrafish. The researchers identify a small set of melanophore progenitors (MPs) that are established early in embryonic development and that are associated with the segmentally reiterated dorsal root ganglia in the fish. They use lineage analysis and four-dimensional in vivo imaging to show that the progeny of these embryonic MPs spread segmentally and give rise to the melanophores that create the adult melanophore stripes. Other experiments indicate that the MPs require zebrafish kit ligand a (kitlga, also known as slk) to function as MSCs, and that MP establishment depends on ErbB signalling during early embryonic development. Based on their results, the researchers propose that dorsal root ganglia provide a niche for MSCs and suggest that Kit signalling might attract and maintain MSCs in this niche.
On p. 1003, Christiane Nüsslein-Volhard and colleagues investigate the embryonic origin of the melanophores that emerge during juvenile development and that contribute to the striking colour patterns of adult zebrafish. The researchers identify a small set of melanophore progenitors (MPs) that are established early in embryonic development and that are associated with the segmentally reiterated dorsal root ganglia in the fish. They use lineage analysis and four-dimensional in vivo imaging to show that the progeny of these embryonic MPs spread segmentally and give rise to the melanophores that create the adult melanophore stripes. Other experiments indicate that the MPs require zebrafish kit ligand a (kitlga, also known as slk) to function as MSCs, and that MP establishment depends on ErbB signalling during early embryonic development. Based on their results, the researchers propose that dorsal root ganglia provide a niche for MSCs and suggest that Kit signalling might attract and maintain MSCs in this niche.
 On p. 996, Thomas O’Reilly-Pol and Stephen Johnson use clonal analysis to investigate which stages of melanocyte regeneration – establishment of MSCs, recruitment of MSCs to produce committed daughter cells, or the proliferation, differentiation and survival of these daughter cells – are affected by Kit signalling deficits; previous work had shown that a reduction in Kit signalling results in dose-dependent reductions of melanocytes during larval regeneration. The researchers show that the reduction in melanocytes in kita mutants is due to a defect in MSC establishment. By contrast, the other stages of melanocyte regeneration are unaffected. Additional analyses indicate that the MSC establishment defect in kita mutants arises from inappropriate differentiation of the MSC lineage, a finding that confirms and extends the results presented by Nüsslein-Volhard and colleagues.
On p. 996, Thomas O’Reilly-Pol and Stephen Johnson use clonal analysis to investigate which stages of melanocyte regeneration – establishment of MSCs, recruitment of MSCs to produce committed daughter cells, or the proliferation, differentiation and survival of these daughter cells – are affected by Kit signalling deficits; previous work had shown that a reduction in Kit signalling results in dose-dependent reductions of melanocytes during larval regeneration. The researchers show that the reduction in melanocytes in kita mutants is due to a defect in MSC establishment. By contrast, the other stages of melanocyte regeneration are unaffected. Additional analyses indicate that the MSC establishment defect in kita mutants arises from inappropriate differentiation of the MSC lineage, a finding that confirms and extends the results presented by Nüsslein-Volhard and colleagues.
PLUS…
Rooting plant development
 In 1993, Liam Dolan, Ben Scheres and colleagues published a paper in Development detailing the anatomical structure of the Arabidopsis root. As part of the Development Classics series, Ben Scheres discusses how this work underpinned subsequent research on root developmental biology and sparked a detailed molecular analysis of how stem cell groups are positioned and maintained in plants. See the Spotlight article on p. 939
In 1993, Liam Dolan, Ben Scheres and colleagues published a paper in Development detailing the anatomical structure of the Arabidopsis root. As part of the Development Classics series, Ben Scheres discusses how this work underpinned subsequent research on root developmental biology and sparked a detailed molecular analysis of how stem cell groups are positioned and maintained in plants. See the Spotlight article on p. 939
Auxin metabolism and homeostasis during plant development
 Auxin plays important roles during the entire life span of a plant. Auxin metabolism is not well understood but recent discoveries, reviewed by Karin Ljung, have started to shed light on the processes that regulate the synthesis and degradation of this important plant hormone. See the Primer article on p. 943
Auxin plays important roles during the entire life span of a plant. Auxin metabolism is not well understood but recent discoveries, reviewed by Karin Ljung, have started to shed light on the processes that regulate the synthesis and degradation of this important plant hormone. See the Primer article on p. 943
Specialized progenitors and regeneration
 Regeneration in planarians requires a population of cells known as neoblasts. Recent data, discussed by Peter Reddien, indicate that some neoblasts express lineage-specific factors during regeneration and in uninjured animals, suggesting that an important early step in planarian regeneration is neoblast specialization. See the Hypothesis article on p. 951
Regeneration in planarians requires a population of cells known as neoblasts. Recent data, discussed by Peter Reddien, indicate that some neoblasts express lineage-specific factors during regeneration and in uninjured animals, suggesting that an important early step in planarian regeneration is neoblast specialization. See the Hypothesis article on p. 951
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...
 Eleven biologists received some unbelievable news this week: They will each receive 3 MILLION dollars from a newly established award. The Breakthrough Prize was founded by Mark Zuckerberg (Facebook) , Sergey Brin (Google) and venture capitalist Yuri Milner, with the goal of supporting research into life extension and curing diseases.
Eleven biologists received some unbelievable news this week: They will each receive 3 MILLION dollars from a newly established award. The Breakthrough Prize was founded by Mark Zuckerberg (Facebook) , Sergey Brin (Google) and venture capitalist Yuri Milner, with the goal of supporting research into life extension and curing diseases.

 (4 votes)
(4 votes)
 (No Ratings Yet)
(No Ratings Yet)


 Fibroblast growth factors (FGFs) are essential for mesoderm induction in vertebrates and for early mesoderm formation in invertebrate chordates. However, functional studies to date do not support a role for FGF signalling in mesoderm induction in other deuterostomes (animals in which the first embryonic opening forms the anus), such as sea urchins. Thus, the ancestral role of FGF signalling during mesoderm specification in deuterostomes is unclear. On
Fibroblast growth factors (FGFs) are essential for mesoderm induction in vertebrates and for early mesoderm formation in invertebrate chordates. However, functional studies to date do not support a role for FGF signalling in mesoderm induction in other deuterostomes (animals in which the first embryonic opening forms the anus), such as sea urchins. Thus, the ancestral role of FGF signalling during mesoderm specification in deuterostomes is unclear. On  In adult stem cell niches, crosstalk between extrinsic cues (such as signals from neighbouring cells) and intrinsic cues (such as chromatin structure) regulates stem cell identity and activity. Now, on
In adult stem cell niches, crosstalk between extrinsic cues (such as signals from neighbouring cells) and intrinsic cues (such as chromatin structure) regulates stem cell identity and activity. Now, on  Members of the Eaf gene family are involved in tumour suppression and in embryogenesis but what are the molecular mechanisms that underlie these activities? Here (
Members of the Eaf gene family are involved in tumour suppression and in embryogenesis but what are the molecular mechanisms that underlie these activities? Here ( During morphogenesis, the elongation of polarised tissues involves cells within epithelial sheets and tubes making and breaking intercellular contacts in an oriented manner. How cells remodel their junctional contacts is poorly understood but growing evidence suggests that localised endocytic trafficking of E-cadherin might modulate cell adhesion. Now, Samantha Warrington and co-workers (
During morphogenesis, the elongation of polarised tissues involves cells within epithelial sheets and tubes making and breaking intercellular contacts in an oriented manner. How cells remodel their junctional contacts is poorly understood but growing evidence suggests that localised endocytic trafficking of E-cadherin might modulate cell adhesion. Now, Samantha Warrington and co-workers ( On
On  In 1993, Liam Dolan, Ben Scheres and colleagues published a paper in Development detailing the anatomical structure of the Arabidopsis root. As part of the Development Classics series, Ben Scheres discusses how this work underpinned subsequent research on root developmental biology and sparked a detailed molecular analysis of how stem cell groups are positioned and maintained in plants. See the Spotlight article on p.
In 1993, Liam Dolan, Ben Scheres and colleagues published a paper in Development detailing the anatomical structure of the Arabidopsis root. As part of the Development Classics series, Ben Scheres discusses how this work underpinned subsequent research on root developmental biology and sparked a detailed molecular analysis of how stem cell groups are positioned and maintained in plants. See the Spotlight article on p.  Auxin plays important roles during the entire life span of a plant. Auxin metabolism is not well understood but recent discoveries, reviewed by Karin Ljung, have started to shed light on the processes that regulate the synthesis and degradation of this important plant hormone. See the Primer article on p.
Auxin plays important roles during the entire life span of a plant. Auxin metabolism is not well understood but recent discoveries, reviewed by Karin Ljung, have started to shed light on the processes that regulate the synthesis and degradation of this important plant hormone. See the Primer article on p.  Regeneration in planarians requires a population of cells known as neoblasts. Recent data, discussed by Peter Reddien, indicate that some neoblasts express lineage-specific factors during regeneration and in uninjured animals, suggesting that an important early step in planarian regeneration is neoblast specialization. See the Hypothesis article on p.
Regeneration in planarians requires a population of cells known as neoblasts. Recent data, discussed by Peter Reddien, indicate that some neoblasts express lineage-specific factors during regeneration and in uninjured animals, suggesting that an important early step in planarian regeneration is neoblast specialization. See the Hypothesis article on p. 
 (7 votes)
(7 votes)