Shaggy hairs and stem cells
Posted by Erin M Campbell, on 10 January 2012
Our intestinal tissue doesn’t need a New Year’s resolution to keep up its amazing productivity. Our intestinal epithelium is replenished at breakneck speed in an assembly line that begins with stem cells. Today’s image is from a recent Development paper that discusses the importance of Notch signaling in stem cell self-renewal and intestinal homeostasis.
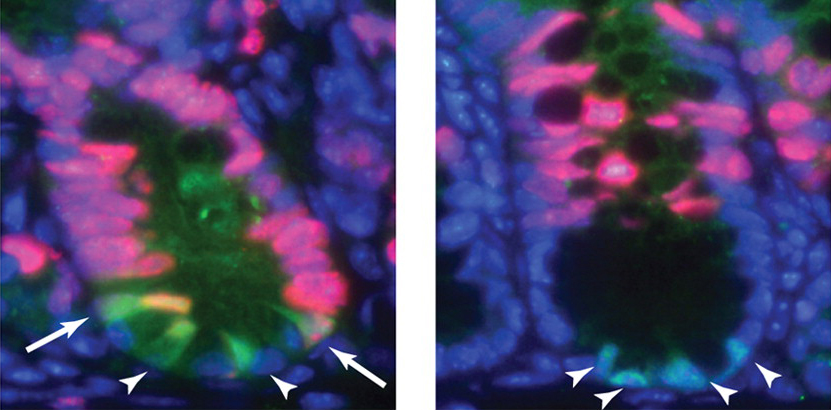
Our intestinal epithelium is folded and shaped into finger-like villi (“shaggy hair” in Latin) that increase the surface area of the tissue for more nutrient absorption. Each villus has several populations of cells in homeostasis in order to maintain function and constant replenishment. This production of epithelium starts with the actively-dividing crypt base columnar (CBC) stem cells that sit in the crypts. Although the identity of these cells has been known for a while, the factors regulating CBC stem cell self-renewal and differentiation were not well understood. A recent Development paper discusses the role for Notch signaling in CBC stem cell function. According to VanDussen and colleagues, Notch signaling is required for CBC stem cell self-renewal and survival. Notch inhibition caused a decrease in the number CBC cells, as well as precocious differentiation of more specialized intestinal cell types. VanDussen and colleagues showed that Notch regulates CBC cell self-renewal and cell fate choice through different pathways and by targeting different cell populations. In the images above, intestinal tissue was stained for a marker of CBC stem cells (Lgr5, green) and for proliferating cells (Ki67, red). In normal tissue (left), CBC stem cells were found at the base of the crypts, some of which were also actively dividing (arrows). Notch inhibition (right) resulted in a misshapen morphology of CBC stem cells, a decrease in the CBC cell marker, and a drop in the number of CBC cells that were actively dividing (arrowheads on left).
For a more general description of this image, see my imaging blog within EuroStemCell, the European stem cell portal.
![]() VanDussen, K., Carulli, A., Keeley, T., Patel, S., Puthoff, B., Magness, S., Tran, I., Maillard, I., Siebel, C., Kolterud, A., Grosse, A., Gumucio, D., Ernst, S., Tsai, Y., Dempsey, P., & Samuelson, L. (2011). Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells Development, 139 (3), 488-497 DOI: 10.1242/dev.070763
VanDussen, K., Carulli, A., Keeley, T., Patel, S., Puthoff, B., Magness, S., Tran, I., Maillard, I., Siebel, C., Kolterud, A., Grosse, A., Gumucio, D., Ernst, S., Tsai, Y., Dempsey, P., & Samuelson, L. (2011). Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells Development, 139 (3), 488-497 DOI: 10.1242/dev.070763


 (3 votes)
(3 votes)
 Somites, confocal, epigenetics, germline, stem cell… BINGO!
Somites, confocal, epigenetics, germline, stem cell… BINGO! Stem Cell Biology
Stem Cell Biology Programme Outline
Programme Outline Why should a developmental biologist be interested in a book about the nucleus? Almost 80 years ago, Conrad Waddington put forward ideas about how gene products could regulate development. In modern parlance, much of development is the result of the differential use of the same genome in different cell types and at different developmental stages within the same organism. This originates in the nucleus, where the processes that act upon the genome – transcription, replication, repair – occur. In developmental biology papers it is not uncommon to find a final summary figure in which a signaling pathway ends up pointing into an oval-shaped nucleus, devoid of any structure or organization, save for a linear depiction of a target gene locus. However, the nucleus is not a homogenous space and neither is the genome in its natural nuclear environment arranged in a linear fashion.
Why should a developmental biologist be interested in a book about the nucleus? Almost 80 years ago, Conrad Waddington put forward ideas about how gene products could regulate development. In modern parlance, much of development is the result of the differential use of the same genome in different cell types and at different developmental stages within the same organism. This originates in the nucleus, where the processes that act upon the genome – transcription, replication, repair – occur. In developmental biology papers it is not uncommon to find a final summary figure in which a signaling pathway ends up pointing into an oval-shaped nucleus, devoid of any structure or organization, save for a linear depiction of a target gene locus. However, the nucleus is not a homogenous space and neither is the genome in its natural nuclear environment arranged in a linear fashion. Ever since Anton van Leeuwenhoek first peered at a living cell in 1674, scientists have been driven to learn everything they can about these tiny units of life and as a result have been developing ever more advanced tools to observe, describe and manipulate them. In the book “The Cell”, a new addition to the Oxford University Press Very Short Introductions series, Terence Allen and Graham Cowling undertook an enormous task of distilling several hundred years of cell biology research into 145 pages including 8 chapters, a further reading section, an index, a glossary and 17 illustrations. The result is that an enormous amount of information is presented in pithy vignettes covering everything from the inner workings of the cell up to the complex interactions of cells within multicellular organisms, as well as cellular disease and directions for future research.
Ever since Anton van Leeuwenhoek first peered at a living cell in 1674, scientists have been driven to learn everything they can about these tiny units of life and as a result have been developing ever more advanced tools to observe, describe and manipulate them. In the book “The Cell”, a new addition to the Oxford University Press Very Short Introductions series, Terence Allen and Graham Cowling undertook an enormous task of distilling several hundred years of cell biology research into 145 pages including 8 chapters, a further reading section, an index, a glossary and 17 illustrations. The result is that an enormous amount of information is presented in pithy vignettes covering everything from the inner workings of the cell up to the complex interactions of cells within multicellular organisms, as well as cellular disease and directions for future research. Benchfly
Benchfly





 (No Ratings Yet)
(No Ratings Yet)