Here are the research highlights from the current issue of Development:
BMP signalling rolls up in the neural tube
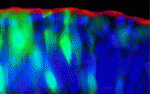
During neurulation, polarised cell-shape changes at hinge points – specialised regions of the neural plate – help convert the neural plate into a tube. But how are these cell-shape changes regulated? To answer this question, Seema Agarwala and co-workers have been studying neural tube closure in the chick midbrain (see p. 3179). They identify a cell cycle-dependent bone morphogenetic protein (BMP) activity gradient in the anterior neural plate and show that it is required for ventral midline hinge point formation and neural tube closure. BMP signalling, they report, regulates the polarised cell behaviours associated with neural tube closure by modulating epithelial apicobasal polarity in tandem with the cell cycle. Because cell-cycle progression in the neural plate is asynchronous, BMP-mediated polarity modulation induces shape changes in only some neural plate cells, whereas their neighbours retain apicobasal polarity. This mosaic and dynamic modulation of polarity, the researchers propose, provides the neural plate with the flexibility to allow folding while retaining its epithelial integrity.
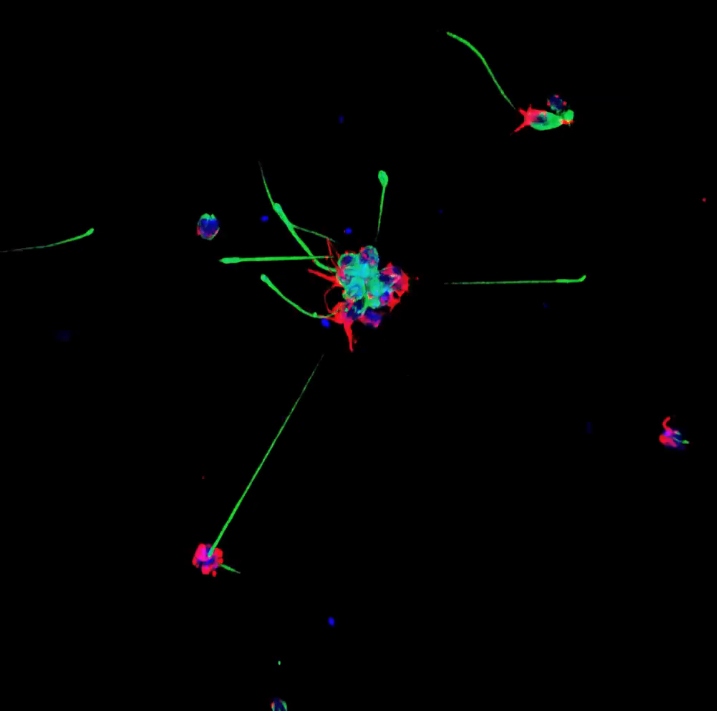
Tetraspanin-like protein regulates islet cell differentiation
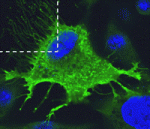
The pancreas is a complex organ that contains ductal, exocrine and endocrine tissues. Here (p. 3213), Kristin Artinger, Lori Sussel and co-workers identify a role for Tm4sf4, a tetraspanin-like protein, during pancreatic endocrine differentiation. Tm4sf4 expression in mice is downregulated by the transcription factor Nkx2.2, which is known to be essential for islet cell differentiation. The researchers show that, in mice, Tm4sf4 is expressed in the pancreatic ductal epithelial compartment and is abundant in islet progenitor cells. Pancreatic tm4sf4 expression and its regulation by Nkx2.2 is conserved in zebrafish, and loss-of-function studies in zebrafish reveal that, in contrast to Nkx2.2, tm4sf4 inhibits α and β cell specification but promotes ε cell fate. Finally, in vitro experiments indicate that Tm4sf4 inhibits Rho-activated cell migration. The researchers propose that the primary role of Nkx2.2 during pancreatic development is to inhibit Tm4sf4 in endocrine progenitor cells, thereby allowing their delamination, migration and differentiation. Targeting Tm4sf4 could, therefore, provide a way to activate quiescent pancreas progenitors for the treatment of diabetes.
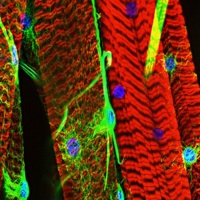
Feel the force: embryonic bone shaping
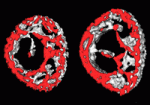
The vertebrate skeleton contains more than 200 bones, each with its own unique shape, size and function. Postnatally, bones remodel in response to the muscle forces they encounter. So bed rest, for example, causes bone thinning. Now, on p. 3247, Amnon Sharir, Elazar Zelzer and colleagues report that muscle force also regulates bone shaping during embryogenesis in mice. Using micro-computed tomography scans of embryonic long bones, the researchers identify a novel developmental programme that, through asymmetric mineral deposition and transient cortical thickening, regulates the specific circumferential shape of each bone. This programme of preferential bone growth, they report, ensures that each bone acquires an optimal load-bearing capacity. Moreover, the programme is regulated by intrauterine muscle contractions; in a mouse strain that lacks such contractions, the bones lose their stereotypical circumferential outline and are mechanically inferior. Thus, the researchers suggest, a reciprocal relationship between structure and mechanical load in utero determines the 3D morphology of developing bones.
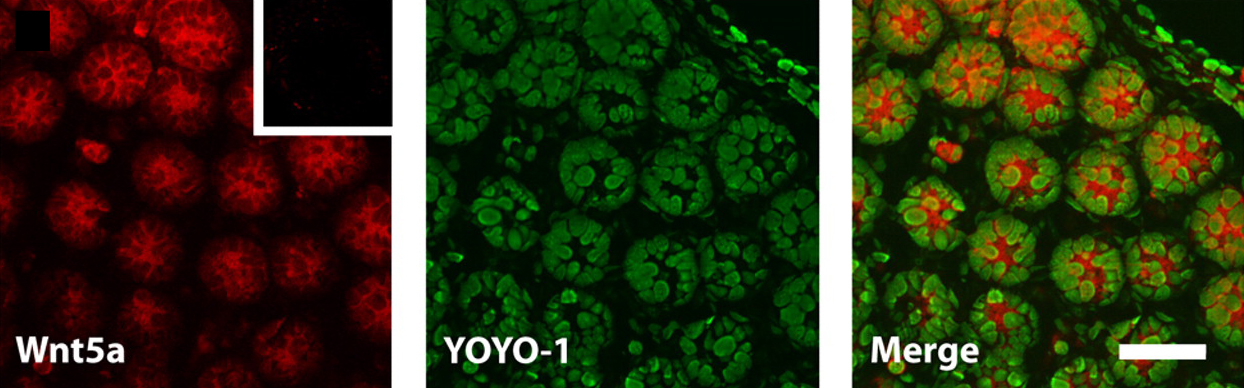
Endoderm specification in sea urchins
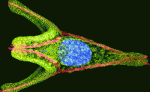
In sea urchin embryos, endomesoderm specification involves β-catenin entry into the nuclei of the vegetal cells of the developing embryo. Now, on p. 3297, David McClay and colleagues reveal how the embryo uses maternal information to initiate this specification by showing that maternal Wnt6 is necessary for activation of endodermal genes. They report that the addition of Wnt6 or ectopic activation of the Wnt pathway rescues endoderm specification in eggs that lack the small region of the vegetal cortex that is normally needed for the activation of the endomesoderm gene regulatory network. This part of the vegetal cortex, they report, contains a high level of Dishevelled (Dsh), a transducer of the canonical Wnt pathway. They also report that morpholino knockdown of Wnt6 in the whole embryos of two sea urchin species prevents endoderm specification but not the expression of mesoderm markers. The researchers suggest, therefore, that maternal Wnt6 plus a localised vegetal cortex molecule, possibly Dsh, are necessary for endoderm specification in sea urchin embryos.
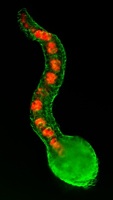
Gata-way to a cardiac progenitor fate
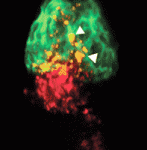
During gastrulation, cardiovascular progenitor cells (CPCs) migrate to the future heart-forming region of the embryo, where they produce the major cardiac lineages. But what regulates CPC fate and behaviour? On p. 3113, Ian Scott and colleagues report that Smarcd3b (Swi/Snf-related matrix-associated actin-dependent regulator of chromatin subfamily d member 3b) and the transcription factor Gata5 can induce a CPC-like state in zebrafish embryos. In mice, SMARCD3, GATA4 and TBX5 form a cardiac BAF (cBAF) chromatin remodelling complex that promotes myocardial differentiation in the embryonic mesoderm. The researchers now show that smarcd3b and gata5 overexpression in zebrafish embryos leads to the formation of an enlarged heart, whereas combined loss of smarcd3b, gata5 and tbx5 inhibits cardiac differentiation. Most notably, transplantation experiments show that cells overexpressing cBAF components migrate to the developing heart and differentiate into cardiac cells, even if initially placed in non-cardiogenic regions of the embryo. These results show that cBAF has a conserved role in cardiac differentiation and can promote a CPC-like state in vivo.
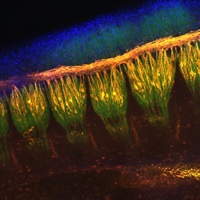
Aired out: FGF9 in lung development
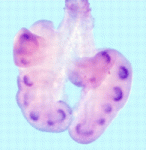
During lung development, the secreted signalling molecule fibroblast growth factor 9 (FGF9) is expressed in both the mesothelium (the single layer of cells that envelopes the lungs) and the pulmonary epithelium (which gives rise to the proximal airways and terminal epithelial buds). Mesenchymal proliferation and epithelial branching are both reduced in Fgf9–/– embryos, which die at birth because of impaired lung development. Intriguingly, David Ornitz and colleagues now show that mesothelial- and epithelial-derived FGF9 have distinct functions during lung development in mouse (see p. 3169). Mesothelial-derived FGF9 and mesenchymal WNT2A, they report, are required to maintain the mesenchymal FGF-WNT/β-catenin signalling pathway that is responsible for mesenchymal growth. By contrast, epithelial-derived FGF9 primarily affects epithelial branching, probably through regulation of BMP4 signalling. The researchers also show that epithelial and mesothelial FGF9 and mesenchymal β-catenin suppress the expression of the BMP4 antagonist Noggin in lung mesenchyme, thereby providing a mechanism for coupling mesenchymal and epithelial proliferation during lung development.
Plus…
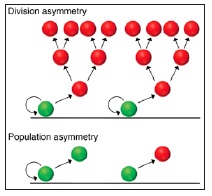
In cycling tissues, stem cells coordinate tissue maintenance and repair. Here, Klein and Simons review the results of recent lineage-tracing studies and challenge the concept of the stem cell as an immortal, slow-cycling, asymmetrically dividing cell.
See the Hypothesis article on p. 3103.
The tenth annual Keystone Symposium on the Mechanism and Biology of Silencing convened in Monterey, California, in March 2011. Olivia Rissland and Eric Lai summarize the results presented at the meeting, which inspire and push this expanding field into new territories.
See the Meeting Review on p. 3093
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (4 votes)
(4 votes)
 (No Ratings Yet)
(No Ratings Yet)







 I am the founder and CEO of DataGiving. I founded DataGiving whilst completing my Ph.D. in Genetics at the University of Cambridge. I have always been passionate about helping people. After completing my Bachelors degree in Psychology, I worked as an Assistant Psychologist at St Marys Hospital in London, helping adults with severe mental health disorders. Since I was a teenager I had aspired to become a Clinical Psychologist, but as much as I admired the great work Psychologists do, I didn’t feel that my desire to reach out and make a positive change would be fully achieved in this role. I returned to academia, as I had long been intrigued to learn more about the biological basis of human behaviour and cognition. I completed a Masters degree in Cognitive Neuroscience, at Imperial College London, which included a laboratory based research project at the Hammersmith Hospital, investigating the genetic basis of Parkinson’s Disease This research sparked my passion for genetics, and specifically the field of Epigenetics. I went on to be awarded an MRC scholarship, to undertake research into the imprinting regulation of Gsα in the laboratory of Dr Gavin Kelsey at The Babraham Institute, Cambridge.
I am the founder and CEO of DataGiving. I founded DataGiving whilst completing my Ph.D. in Genetics at the University of Cambridge. I have always been passionate about helping people. After completing my Bachelors degree in Psychology, I worked as an Assistant Psychologist at St Marys Hospital in London, helping adults with severe mental health disorders. Since I was a teenager I had aspired to become a Clinical Psychologist, but as much as I admired the great work Psychologists do, I didn’t feel that my desire to reach out and make a positive change would be fully achieved in this role. I returned to academia, as I had long been intrigued to learn more about the biological basis of human behaviour and cognition. I completed a Masters degree in Cognitive Neuroscience, at Imperial College London, which included a laboratory based research project at the Hammersmith Hospital, investigating the genetic basis of Parkinson’s Disease This research sparked my passion for genetics, and specifically the field of Epigenetics. I went on to be awarded an MRC scholarship, to undertake research into the imprinting regulation of Gsα in the laboratory of Dr Gavin Kelsey at The Babraham Institute, Cambridge.