In our recently published paper1, we show that human stem cells self-organize into blastocyst-like structures, which we term blastoids based on 4 criteria. Because blastoids can be generated in large numbers, can be finely manipulated and recapitulate aspects of endometrial implantation in vitro, they are powerful models to investigate the principles of blastocyst development and implantation. Here I want to tell the backstory of our scientific journey and why I believe it is important for science and medicine.
Nature | DOI: 10.1038/s41586-021-04267-8
Human embryology
Understanding human development is one of the new frontiers in developmental biology. Because of the intrinsic difficulties in studying human embryos, the majority of our knowledge has been inferred from model organisms such as mice, frogs, fish, or fruit flies. However, many developmental principles are not conserved and humans have evolved specific features2. Human embryos can be studied when generously donated by couples undergoing an IVF procedure, and this has proved extremely useful3. However, IVF embryos are scarce, not always of top quality, and considerably difficult to work with. On the contrary, embryo models formed from pluripotent stem cells (hPSCs), although they are simplified versions of the embryo, can be formed easily from established cell lines in the lab, generated in large numbers, and are easier to manipulate either physically or genetically. They thus represent an ethical and technical alternative complementing the use of IVF embryos.
Understanding implantation with blastoids
In our lab (Nicolas Rivron, IMBA-Institute for Molecular Biotechnology, Austrian Academy of Sciences), we are interested in understanding early mammalian development and how the embryo implants into the uterus. We are especially studying the formation of the blastocyst (day 4-7 in humans) that generates and pattern tissues to prepare the conceptus for implantation into the uterus. Implantation is the first physical contact between the mother and the embryo and an extremely crucial step towards the sustenance of the pregnancy. Implantation success marks the establishment of a clinical pregnancy as measured by levels of the hormone human chorionic gonadotropin. Of clinical importance, in humans, the steps of blastocyst development and implantation are prone to failure and are thought to account for as much as 75% of pregnancy loss4,5.
Despite its tremendous significance, understanding blastocyst development and implantation has proven to be a challenging endeavor. This is because embryos are available in limited numbers and their interactions with the uterus happens deep inside the womb, therefore is inaccessible and invisible. These limitations in working with IVF embryos and the uterus restrain the possibilities to perform, for example, complex genetic manipulations, biochemistry, as well as high throughput chemical or genetic screens that are at the heart of scientific and biomedical discoveries. To overcome these challenges, our lab has previously developed a model of mouse blastocyst generated solely using stem cells, which we termed blastoid6. Mouse blastoids formed through the combination of cells from lines of mouse embryonic stem cells7 and mouse trophoblast stem cells8. When treated with a defined cocktail of growth factors and/or their modulators, these two stem cell types interact, which triggers their self-organization into structures that are remarkably similar to mouse blastocysts6,9,10. The mouse blastoids was a first complete model of the conceptus and, because it modeled the pre-implantation stage, we were able to transfer them in utero to study implantation. No mice were formed from mouse blastoids until now but blastoids are a scalable and experimentally amenable platform to employ multimodal approaches such as complex genetic manipulations and high-throughput imaging in order to address scientific questions such as principles of embryonic/extraembryonic inductions and self-organization.
2021: Many attempts to model human blastocysts.
The mouse blastoids reported in 2018 triggered interest to form human blastoids and the potential clinical implications multiplied that enthusiasm. Till date, 6 groups independently proposed human blastocyst models, all of which were published in 2021. These 6 methods were developed using at least 4 different states of stem cells as a starting point, and 6 different culture conditions to tentatively trigger the formation of analogs of blastocyst lineages. The output with respect to the stage of the cells that were generated, the level of adequacy with developmental time and sequence, and the efficiency of the formation of structure widely varied. Overall, establishing ways of evaluating the weight of the initial experimental parameters and of defining criteria to assess the cells and structures that are formed will be essential. To our best of understanding, the initial cell state and the combination of molecules used to trigger blastoid formation are the two main parameters that, when combined, allow for modeling blastocyst development and forming the right cells reflecting the blastocyst stage.
Currently, the most powerful way to perform end-point assessments is to combine single cell RNA sequencing (scRNA seq) data of the model with a reference map generated from various staged human embryos (pre-blastocyst, blastocyst, post-implantation stages). Having multiple stages, including the ones before and after the blastocyst stage, is essential to detect potential off-target cells. Fortunately, the last few years have been fruitful in generating high-quality datasets from human embryos including IVF pre-blastocyst and blastocysts11,12, IVF blastocysts further cultured in vitro for up to 14 days13, that mimic aspects of early post-implantation development. Moreover, a very recent study has created an invaluable scSeq dataset from a gastrulation human embryo (Carnegie stage 7)14. Overall, this provides an unprecedented opportunity to generate a high-quality reference meta-map from human embryos in order to evaluate embryo models. In the future, additional layers of readouts (e.g., ATAC seq, Bi-sulfite seq, etc.) would be useful to complement the analysis of the transcriptome alone.
Our own attempt to form human blastoids.
In our study1, we report the generation of human blastoids that, to the best of our knowledge, faithfully and efficiently recapitulate key features of blastocyst development: the sequence and the pace of lineage specification and morphogenesis, which results in the formation of blastocyst-like cells. We measured 97% of blastocyst-like cells based on criterias that we think should be challenged, fine-tuned, and complemented. This reliability in modeling blastocyst development allowed us to subsequently model several aspects of implantation using endometrial organoids. Here we describe the initial cell state and combination of molecules triggering the formation of blastoids, and the end-point criteria that we think are valid to evaluate such models. the human blastocyst.
The right shade of pluripotent stem cells: a secret ingredient to faithful blastoids
In mice, the culture conditions to establish stem cell lines reflective of the blastocyst lineages are relatively well established. For example, stem cells can be captured in a state that reflects the blastocyst-stage epiblast, and that are referred to as 2i/Lif naive PSCs7. Similarly, the blastocyst-stage trophectoderm can be captured in a state that we refer to as trophectoderm stem cells (TESCs, unpublished manuscript)9. This allows these two developmentally matched stem cell types to efficiently self-organize and to form mouse blastoids. Altogether, to form a faithful and thus useful blastoid, it is crucial to begin with stem cells that reflect the blastocyst stage, and that are capable of efficiently forming and organizing analogs of the blastocyst lineages upon exposure to developmentally relevant cues.
In mice, the culture conditions to establish stem cell lines reflective of the blastocyst lineages are relatively well established. For example, stem cells can be captured in a state that reflects the blastocyst-stage epiblast, and that are referred to as 2i/Lif naive Pluripotent Stem Cells (PSCs)7. Similarly, the blastocyst-stage mouse trophectoderm can be captured in a constrained state that we refer to as trophectoderm stem cells (TESCs)9[now published in Cell Stem Cell]. This allows these two developmentally matched stem cell types to efficiently self-organize into mouse blastoids. Altogether, to form a faithful and thus useful blastoid, it is crucial to begin with stem cells that reflect the earliest stage possible, and that are capable of efficiently forming and organizing analogs of the blastocyst lineages upon exposure to developmentally relevant cues.
The human stem cells that best reflect the blastocyst stage have been difficult to capture. The firstly established and most commonly used hPSCs15 are referred to as primed hPSCs and are transcriptionally similar to the epiblast of post-implantation staged embryos (day 12-14) as compared to blastocysts (day 5-7)16. This is typically shown by measuring stage-specific molecular criteria (e.g., genome-wide DNA hypomethylation, X chromosome status17,18) and by comparing the cells transcriptome with reference monkey19 or human datasets11,13,14. However, in the last decade, multiple labs have shown that primed hPSCs cultured with specific signaling pathway inhibitors and activators are capable of resetting their transcriptome, decreasing the methylation of their DNA, and, to some extent, to reactivate silenced X chromosome17,18,20–22. These shades of naive pluripotency reflect the blastocyst epiblast (~ day 6). Other attempts to reset primed hPSCs allowed to capture other states, for example human extended potential stem cells (hEPSCs)23 but, upon re-analysis, these cells display a transcriptome more similar to the post-implantation stage (see analysis in Figure 724 and Figure 325).
The unrestricted potential of naive human pluripotent stem cells
The mouse and human blastocysts comprise 3 lineages: the trophectoderm that mediates implantation and then generates the placenta, the epiblast that forms the majority of the body, and the primitive endoderm that forms the yolk sac and part of the embryonic endoderm. In mice, the specification of the trophectoderm occurs during the transition from a morula to a blastocyst26. On the contrary, studies by the laboratories of Laurent David (INSERM and Université de Nantes, France) and Hilde van de Velde (Vrij Universiteit Brussel) have suggested that the early human blastocyst doesn’t comprise fully specified cells but rather cells that are not yet entirely restricted to a specific lineage12 27. In correlation with this observation, the laboratories of Yasuhiro Takashima (CiRA) and of Austin Smith (Exeter University) have shown that naive hPSCs cultured in either PXGL20 or Ti2iLGö18 are capable of forming analogs of the blastocyst-stage trophectoderm28,29. Once again, this potential is in sharp contrast to the already determined potential of mouse PSCs that cannot efficiently form trophoblasts, as assessed in chimeric and in vitro assays30–32. Of note, although several studies have described the potential of primed hPSCs or hEPSCs to differentiate into cells with trophoblast-like properties, this process does not seem efficient, and the obtained cellular populations might be heterogeneous and reflecting a mix of post-implantation trophoblasts and amnion cells (a lineage thought to emerge during the post-implantation period) rather than the trophectoderm (that forms in the blastocysts). However, these attempts to efficiently form trophoblasts from primed hPSCs were very successful in order to learn about the signaling pathways33,34 regulating aspects of trophoblast specification and pinpointing the important plasticity of human cells as compared to mouse cells.
The right signalling activity combination- TGF-β, ERK and Hippo inhibition: other secret ingredient
Therefore, considering the similarities of naive PXGL hPSCs to the blastocyst epiblast and the enhanced plasticity of human cells as compared to mice, we tested the potential of PXGL hPSCs to form human blastocyst-like structures. To do so, we formed a small aggregate of cells (~80-100 um) and provided molecules that were previously found to support either the formation of human trophoblasts (inhibition of TGF-β with A83-0133, ERK with PD0328,29,33,35) or to participate in the maintenance of mouse TESCs (inhibition of Hippo pathways with LPA9), and to maintain naive hPSCs (LIF)21, along with a ROCK inhibitor to enhance cell survival and aggregation. To our surprise, within 96 hours, this culture condition was sufficient to induce the very efficient self-organization of the aggregates into structures that bear remarkable morphological resemblance to the human blastocyst. This protocol is robust and, with minor tuning of concentration of inhibitors, multiple hPSC,both ESC and iPSC, lines formed such structures with remarkably high efficiency (70-90% for every cell line we tested).
Fantastic 4 features: Rise of the human blastoids
For embryo models to be valuable, they must possess a good level of analogy with the embryo. Therefore, here we benchmarked our blastocyst-like structures using several known features of the human blastocyst. Here, we propose 4 cardinal features that define a human blastoid.
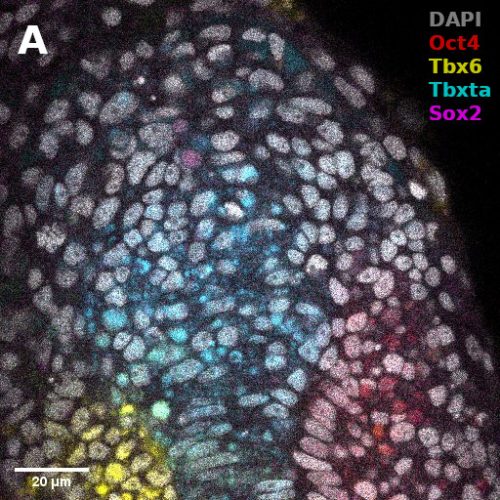
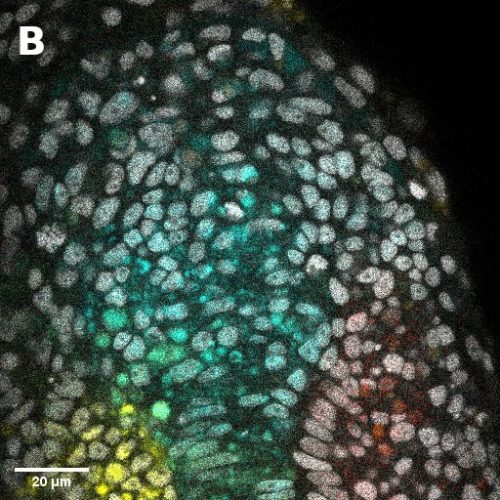
1. Size, morphology, and cell numbers: A fully expanded human blastocyst consists of a trophectoderm cyst of diameter between 150-250 μm that surrounds a single fluid-filled cavity and encompasses a single cluster of inner cells. We used the threshold of 150-250um diameter to define a blastoid and thus their yield of formation. These blastoids showed remarkable morphological resemblance to the human blastocysts and surprisingly, almost always form only one inner cell cluster of Inner Cell Mass (ICM)-like cells. Moreover, the total number of cells as well as ratios between various compartments are conserved as well (Figure 1h1). However, in order to better benchmark blastocyst-like structures, a more detailed and more extensive reference dataset of cell numbers within each lineages and over blastocyst development time (e.g., day 4-7), would be highly valuable.
2. Timing and sequence of development: The human blastocyst forms towards the end of the 4th day after fertilization and continues to develop for the next 3 to 4 days, at which time (~days 7-8) it undergoes implantation. Thus, blastocyst development from its formation to implantation takes 3 to 4 days. During that time, a precise sequence of events happens: First, the trophectoderm and ICM form. Then, in a second step, the ICM generates GATA4+ primitive endoderm cells and the trophectoderm matures to form NR2F2+ polar trophectoderm. This developmental dynamics of lineage specification was spontaneously recapitulated during blastoid formation: Similar to human blastocysts, trophectoderm and ICM analogs were the first lineages to form (24-60 hours), while primitive endoderm and polar trophectoderm analogs only formed in the second stage (60-96 hours). Of note, analogs of primitive endoderm and of the polar trophectoderm formed autonomously, solely based on the initial conditions, and probably due to intrinsic cell interactions and positional information originating from the overall geometry of the structures. No external stimulations are necessary during intermediate stages.. This pace matching is important in order to ultimately form cells that reflect the blastocyst stage. The necessity to culture cells for a longer time in order for morphogenesis to occur properly would also lead to the formation of cells reflecting the post-implantation rather than pre-implantation stage.
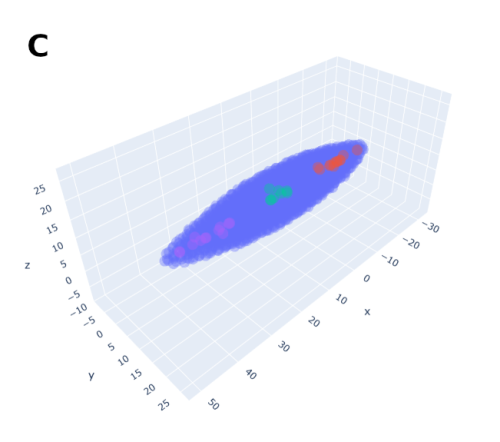
3. Transcriptomic states and stage: Human blastocysts ultimately consist of only three cell types, epiblast, trophectoderm and primitive endoderm that are each characterized by a distinct transcriptome. Importantly, these transcriptomes significantly differ from that of their derivatives in the post-implantation stages. It is thus possible to qualitatively infer the stage equivalent of the cells based on their transcriptome. However, methods to put hard numbers on this remain insufficiently developed. It is crucial for blastoids to be composed of analogs of only the blastocyst cell types and devoid of their post-implantation derivatives or of any other ‘off-target’ cell types. Several recent studies have reported high quality single cell transcriptomic datasets from human embryos of various stages ranging from morula to gastrula11,13,14. The latest one was produced by the laboratory of Laurent David (INSERM and Université de Nantes, France) who combined a metadataset and developed a pseudotime trajectory that appeared extremely useful36. An efficient and relaxed collaboration with this laboratory allowed us to have precise reference maps in order to determine the stage of the cells formed within the blastocyst-like structures and the precise cell states composing them. Ultimately, the blastocyst-like structures formed contained only three main, transcriptionally distinct clusters and each individual cluster showed a gene signature of epiblast, trophectoderm and primitive endoderm respectively. However, the decisive experiment came when we projected these cells on the human embryonic reference map consisting of cells isolated from pre-blastocyst, blastocyst, in vitro cultured blastocyst (equivalent of day 7 to day 14), and gastrulating embryos (CS7). The cells isolated from the blastocyst-like structures coincided quite precisely with the transcriptome of the cells of the blastocyst but not with pre-blastocyst stages or any post-implantation stages. Specifically, the transcriptome of the cells that formed overtime matched with day 5 blastocysts after 24 hours, and with day 7 blastocysts after 96 hours thus revealing the stages that are modeled. We found a minor population of ‘off-target’ cells which accounted for less than 3% of the cells analyzed. Thus, with morphological resemblance to human blastocyst, efficient formation of analogs of blastocyst cells according to the sequence and pace of blastocyst, and absence of off-target cells, we now felt confident to refer to these structures as blastoids.
4. Implantation and post-implantation developmental potential: Between day 4 and day 7 after fertilization, the blastocyst forms a cavity, which expands. During this, the blastocyst prepares itself to interact with the uterus so as to implant in the right direction and facilitate subsequent embryonic development. For example, the trophectoderm patterns into 2 distinct zones: the trophectoderm cells closer to the epiblast form the polar trophectoderm while the cells on the opposite end form the mural trophectoderm. In human blastocysts, such patterning is key for implantation as the polar trophectoderm forms the sticky cells that mediate the initial interactions with the endometrial cells of the uterus. Strikingly the trophectoderm analogs of the majority of blastoids (>60%) were patterned into polar and mural-like trophectoderm cells as seen by their protein expression profile (NR2F2High/CDX2Low). This indicated that the blastoids may possess the ability to undergo directional implantation. However, it is of course unethical and now forbidden by the ISSCR to transfer human blastoids into the uterus of humans or any other model organisms for any purpose including for scientific experiments37,38.
Therefore, we developed, through a fruitful and fun collaboration with Nina Maenhoudt and Hugo Vankelekom (KU Leuven, Belgium), an in vitro implantation assay using the cells from the endometrial organoids. We were extremely fascinated to see that, when blastoids were deposited on endometrial cells previously stimulated with hormones, blastoids attached to the endometrial cells thus mimicking the first step of implantation. However, this interaction did not occur if endoTherefore, we developed, through a fruitful and fun collaboration with Nina Maenhoudt and Hugo Vankelekom (KU Leuven, Belgium), an in vitro implantation assay using the cells from the endometrial organoids. We were extremely fascinated to see that, when blastoids were deposited on endometrial cells previously stimulated with hormones, they attached to the endometrial cells thus mimicking the first step of implantation. However, this interaction did not occur if endometrial cells were not stimulated with hormones. In accordance with clinical knowledge, we concluded that the hormones changed the state of the endometrial cells and made them receptive for the blastoid. Even more surprisingly, upon live imaging, we realized that this initial interaction was reproducibly mediated by the cells on the polar side of the trophectoderm. This was an exciting moment for us because replicating this directional attachment to hormonally stimulated endometrial cells gives confidence that we can mimic important aspects of the initiation of implantation. Importantly, the exposure of blastoids to a STAT inhibitor (SC144) and to a strong Hippo inhibitor (XMU-MP01) prevented the formation/maintenance of the inner cell cluster and thus to the formation of trophospheres. These trophospheres that transcriptionally appear to most resemble mid-stage blastocysts were incapable of attaching to hormonally stimulated endometrial cells. These additional controls reinforced our conviction that this assay demonstrated the specificity and reliability of blastoids to mimic aspects of implantation. It also opens up new possibilities to understand the molecular basis of embryo-uterus cross talk during implantation. Finally, blastoids can also be cultured for an extended period of time after their attachment to endometrial cells, during which they consistently expand and attain several features of post-implantation human embryos. For example, some trophectoderm cells differentiate into syncytiotrophoblast and extravillous trophoblast; the epiblast begins to express CD24 and appear more epithelial. However, since the methods to culture human blastocysts to post implantation stages are currently suboptimal, the extended blastoid cultures cannot be appropriately benchmarked. We do not think that we have properly modeled a day 13 stage. Nevertheless, blastoids cultured to extended periods augurs well for using these to study peri-implantation embryonic development.
The above criteria canThe above criteria can be used to determine the reliability and predictive power of any in vitro blastocyst model. We propose that these initial criteria must be thoroughly evaluated in order to define the in vitro structures as blastoids. However, we also acknowledge that these criteria are currently minimal. With further research more features such as the proteomic and epigenetic status, the mechanisms of lineage specification, the X chromosome status, etc., must be benchmarked to the human blastocysts and such features measured to evaluate the quality and utility of the model.
Publishing human blastoids: A long roller coaster ride
The journey between the generation of the first human blastoid in our lab to the first submission of our manuscript was an intense one. Several members of the team took responsibility for different aspects of the study and worked in a highly coordinated manner. Though intense, the team made this journey an exciting and delightful one. We submitted our manuscript with a completely optimized blastoid to the journal Nature on the 12th of February 2021. At that time, no other protocol to form models of the human blastocyst had been published. The editor received our manuscript with a lot of enthusiasm but there was definitely a shared sense of urgency to publish it. As a consequence, the first round of the peer review process was accelerated as well. We are extremely thankful to our reviewers Jan Bronsens, Jianping Fu, Insoo Hyun (open reviewers) and 2 other anonymous reviewers for the fast yet mostly constructive criticisms and the suggested experiments. However, the editor gave us a hard deadline of only 4 months to revise the manuscript and only one chance for re-submitting a manuscript that would address all the reviewer’s comments.
This brought back the intense phase, but this time with a deadline. Our synergistic work and the help of Maria Novatchkova (an excellent bioinformatician at the Vienna Biocenter) and collaborators, especially in that revision phase, from the lab of Laurent David, made it possible to submit a revised manuscript after approximately 3 months, about one month before the deadline. However, during this phase several reports were published reporting blastocyst-like structures, albeit with low efficiency. Though we were disappointed, we were rejoiced by the interest that the scientific community has towards this field.
Within weeks after the first attempts were released, a consortium of independent stem cell biologists and embryologists led by Fredrik Lanner (Karolinska Institute) and Sophie Petropoulos (Montreal University) gathered to evaluate the state and the stage of the cells within these models. They used an extensive reference map formed by parametrizing and merging multiple datasets of cells harvested a pre-blastocyst, blastocyst, and post-implantation stages. The initial report from this consortium was very rapidly published on Biorxiv and reported the abundance of ‘non-blastocyst’ like cells that closely resemble several post-implantation cell types present in the amnion, mesoderm, and primitive streak (~day 14)39. The presence of such ‘off-target’ cells is probably due to the sub-optimal initial cell state and/or suboptimal combination of molecules, which results in slow development (6 days or 9 days) and in a process of directed differentiation that led to the formation of cells similar to the gastrulation (day 14) and germ layer (mesoderm) stages. We suspect this, along with the low efficiency, can greatly reduce the utility of the model. It reignited our enthusiasm to finalize our proposition to form a model of human blastocyst-stage embryo.
While setting up rapid and good standards for the field, this independent consortium report probably caused an unprecedented scrutiny over our manuscript. We received very positive comments from 2 out of 3 of the reviewers which hinted our optimistic minds towards the acceptance of our manuscript. In the meantime, an ethicist, who turned out to be Insoo Hyun (open reviewer no. 4) gave us an ethical green light based on compliance with ISSCR guidelines and on the assessed oversight by the Austrian Academy of Sciences. However, the editor decided to add an additional reviewer (Reviewer no. 5) specifically to ensure the validity of the generation and analysis of the single cell transcriptomic data used to claim the formation of cells reflecting the blastocyst stage. Although disappointed by that unexpected move that consumed more time than we anticipated, we were excited to learn a bit more than one month later, at the end of August, that our scRNA seq analyses had passed this additional test. In addition, an update of the meta-analysis led by Fredrik Lanner and Sophie Petropoulos also included our model along with the one formed in the lab of Ge Guo (Exeter University) and showed that these models reflect the blastocyst stage. Forming the right cells is crucial because only faithful embryo models will be capable of reliably predicting human development. This will ensure that we can trustfully use embryo models to infer the (epi)genetic mechanisms, molecular pathways and cellular behaviors of blastocysts. While this first report from the consortium is important, more detailed analysis (such as quantitative assessment of the resemblance of a specific cell type of a blastocyst model to the one of the blastocyst) would be extremely beneficial to assess degrees of similarity. Such an understanding of the initial cell state and of the combination of molecules that allow for the formation of human blastoids also call for unbiased re-analysis of single cell RNA sequencing (scSeq) by independent bioinformaticians. It would support valid scientific and medical discoveries, and assist ethical evaluations.
The editor then sent us a ‘provisional’ acceptance (provided it clears the editorial criteria) on September 3rd, 2021. However, for reasons that we do not understand, we had to anxiously wait for more than 75 days for our manuscript to become formally accepted on the 18th of November, 2021. Finally, the study was published on the 2nd of December 2021, 3 months after provisional acceptance, through an accelerated track gladly proposed by the editor.
We hope that the scientific community has read our study with as much excitement as we had when first realizing that these blastoids form reliably according to the cardinal features of human blastocysts (size/morphology/cell numbers, timing/sequence of development, transcriptomic states/stage, functional implantation, and post-implantation developmental potential). Our data points to the possibility that the proposed blastoid is valuable to study early human development. Overall, this scientific journey shows that the initial cell state (PXGL naïve hPSCs) and the combination of molecules (triple inhibition of the Hippo/ERK/TGF-β pathways) allow for the formation of human blastoids that accurately form blastocyst-stage cells. However, we strongly encourage independent groups and bioinformaticians to critically work with our sequencing data sets to compare with that of human embryos to verify or challenge our analysis. Afterall, self-correction can only allow for the field to progress toward more stringent criteria and models predictive of human blastocyst development and implantation.
Blastoid team: great companions in a gratifying journey
Working on the project to develop human blastoids was a rare opportunity that I was extremely privileged to experience. I am extremely thankful to my supervisor Nicolas Rivron for providing me this opportunity. The most striking aspect to me was his broad vision behind the development of the project and the tremendous planning to execute the ideas in a highly coordinated manner. This was made possible by the extremely supportive and helpful atmosphere in the lab. When I started working in this lab, in January 2020, I joined the efforts of an extremely talented postdoc, Harunobu Kagawa, in characterizing naive PSCs and human trophoblast stem cells. Meanwhile, in parallel, another postdoc, Heider Hedari Khoei, who joined the lab just 2 days after me, used his clinical expertise on endometrial organoids. Through collaborative work with Nina Maenhoudt and Hugo Vankelekom (KU Leuven, Belgium), he developed the in vitro implantation assay. Thus, when we successfully developed our first human blastoid, we had already established a system to assess their functionality by the implantation assay. This synergistic team work within the lab (Theresa Sommer and Yvonne Scholte Op Reimer performed several crucial experiments and Giovanni Sestini led all the analysis of all the sequencing data) as well as collaborations (Labs of Laurent David, Hugo Vankelekom and Thomas Freour) allowed for the realization of this exciting study. This has been a learning experience and a journey that I will cherish. Blastoids have now opened up a new opportunity to dive deep into exploring mechanisms underlying early embryogenesis in humans. Thus, the exciting journey continues.
References
1. Kagawa, H. et al. Human blastoids model blastocyst development and implantation. Nature (2021) doi:10.1038/s41586-021-04267-8.
2. Gerri, C., Menchero, S., Mahadevaiah, S. K., Turner, J. M. A. & Niakan, K. K. Human Embryogenesis: A Comparative Perspective. Annu. Rev. Cell Dev. Biol. 36, 411–440 (2020).
3. Gerri, C. et al. Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature 587, 443–447 (2020).
4. Wilcox, A. J., Baird, D. D. & Weinberg, C. R. Time of implantation of the conceptus and loss of pregnancy. N. Engl. J. Med. 340, 1796–1799 (1999).
5. Norwitz, E. R., Schust, D. J. & Fisher, S. J. Implantation and the survival of early pregnancy. N. Engl. J. Med. 345, 1400–1408 (2001).
6. Rivron, N. C. et al. Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111 (2018).
7. Ying, Q.-L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
8. Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A. & Rossant, J. Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 (1998).
9. Frias-Aldeguer, J. et al. Embryonic signals perpetuate polar-like trophoblast stem cells and pattern the blastocyst axis. Preprint at https://doi.org/10.1101/510362.
10. Vrij, E. J. et al. Chemically-defined induction of a primitive endoderm and epiblast-like niche supports post-implantation progression from blastoids. Preprint at https://doi.org/10.1101/510396.
11. Petropoulos, S. et al. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 165, 1012–1026 (2016).
12. Meistermann, D. et al. Spatio-temporal analysis of human preimplantation development reveals dynamics of epiblast and trophectoderm. Preprint at https://doi.org/10.1101/604751.
13. Zhou, F. et al. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature 572, 660–664 (2019).
14. Tyser, R. C. V. et al. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature (2021) doi:10.1038/s41586-021-04158-y.
15. Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
16. Nichols, J. & Smith, A. Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 (2009).
17. Theunissen, T. W. et al. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 19, 502–515 (2016).
18. Takashima, Y. et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269 (2014).
19. Nakamura, T. et al. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature vol. 537 57–62 Preprint at https://doi.org/10.1038/nature19096 (2016).
20. Guo, G. et al. Epigenetic resetting of human pluripotency. Development 144, 2748–2763 (2017).
21. Chen, H. et al. Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nat. Commun. 6, 7095 (2015).
22. Gafni, O. et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286 (2013).
23. Yang, Y. et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 169, 243–257.e25 (2017).
24. Stirparo, G. G. et al. Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast. Development 145, (2018).
25. Castel, G. et al. Induction of Human Trophoblast Stem Cells from Somatic Cells and Pluripotent Stem Cells. Cell Rep. 33, 108419 (2020).
26. Rossant, J. Making the Mouse Blastocyst: Past, Present, and Future. Curr. Top. Dev. Biol. 117, 275–288 (2016).
27. De Paepe, C. et al. Human trophectoderm cells are not yet committed. Hum. Reprod. 28, 740–749 (2012).
28. Guo, G. et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 28, 1040–1056.e6 (2021).
29. Io, S. et al. Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell 28, 1023–1039.e13 (2021).
30. Posfai, E. et al. Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 23, 49–60 (2021).
31. Baker, C. L. & Pera, M. F. Capturing totipotent stem cells. Cell Stem Cell 22, 25–34 (2018).
32. Morgani, S., Nichols, J. & Hadjantonakis, A.-K. The many faces of Pluripotency: in vitro adaptations of a continuum of in vivo states. BMC Dev. Biol. 17, 7 (2017).
33. Amita, M. et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc. Natl. Acad. Sci. U. S. A. 110, E1212–21 (2013).
34. Xu, R.-H. et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264 (2002).
35. Daoud, G. et al. ERK1/2 and p38 regulate trophoblasts differentiation in human term placenta. J. Physiol. 566, 409–423 (2005).
36. Meistermann, D. et al. Integrated pseudotime analysis of human pre-implantation embryo single-cell transcriptomes reveals the dynamics of lineage specification. Cell Stem Cell 28, 1625–1640.e6 (2021).
37. Lovell-Badge, R. et al. ISSCR Guidelines for Stem Cell Research and Clinical Translation: The 2021 update. Stem Cell Reports 16, 1398–1408 (2021).
38. Clark, A. T. et al. Human embryo research, stem cell-derived embryo models and in vitro gametogenesis: Considerations leading to the revised ISSCR guidelines. Stem Cell Reports 16, 1416–1424 (2021).
39. Zhao, C. et al. Reprogrammed iBlastoids contain amnion-like cells but not trophectoderm. bioRxiv (2021) doi:10.1101/2021.05.07.442980.
 (13 votes)
(13 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet)
 (1 votes)
(1 votes)