MCC loss during mucociliary epithelium remodeling: new insight into a debated topic and a decade mystery.
Posted by Alexia Tasca, on 29 March 2021
Introduction
In mucociliary epithelia, such as the mammalian airway epithelium or the embryonic epidermis of Xenopus tadpoles, the correct balance between multiciliated cells (MCCs) and secretory cells provides the functional basis for removal of particles and pathogens to prevent infections and to maintain organismal oxygenation (Walentek and Quigley, 2017).
While the rest of the Walentek lab is working on how mucociliary epithelia are established during development, I am the “black sheep” of the lab, because I wanted to know how this tissue is remodeled to become a non-ciliated epithelium during metamorphosis. Indeed, mucociliary epithelial remodeling and MCC loss are observed in human chronic lung disease as well as during metamorphosis of the Xenopus epidermis. However, it remained unresolved how and why MCCs were lost in Xenopus, and how the process compares to observations made in mouse models of airway inflammation and human cells from chronic airway disease patients. By addressing this question, we hoped to find the underlying molecular mechanism for MCC loss in Xenopus, and to establish a new model to study mucociliary remodeling in the vertebrates.
Thus, I started my PhD on the loss of MCCs during Xenopus tadpole development. I found it especially interesting that everyone in the field was aware of this loss, but no one really looked at it in detail before to see how and when this was precisely happening. The first paper that I found on this topic dated from 1988 (Smith et al, 1988), where the authors describe a loss of MCCs from areas around the developing lateral line. Additionally, another group described MCCs with reduced ciliation that were positive for mucus staining in advanced tadpole stages (Nishikawa et al., 1992). But then, I could not find further work that would explain these phenomena. It remained unclear how the complete loss of MCCs from the tadpole epidermis was accomplished and why MCCs were lost in the first place. Therefore, I got interested in the case and I was hoping to resolve this mystery like a detective.
At the start of my investigations, I first established the time line of MCC loss during Xenopus laevis tadpole development by immunofluorescence confocal- and scanning electron-microscopy.Being new to the field of Xenopus epidermis biology, my first discovery was to see how amazing normal MCCs look, with their hundreds of motile cilia and their dense apical F-actin network. Then, I started to observe how their morphology changed over time of epidermal remodeling. During these studies, I found that MCCs were lost during a first “local” phase in areas where lateral line neuromasts (NMs) would emerge. A bit later, MCCs were also lost everywhere else in the epidermis. This suggested to me that there could be two distinct mechanisms for MCC loss, depending on the location and the timing. So, I set out to investigate both processes in more detail.
MCCs undergo lateral line-induced apoptosis
Investigating the relationship between MCC loss and neuromast (NM) development seemed like a good start, because it confirmed previous findings by Smith et al. in other frog species and demonstrated a conservation of this phenomenon.
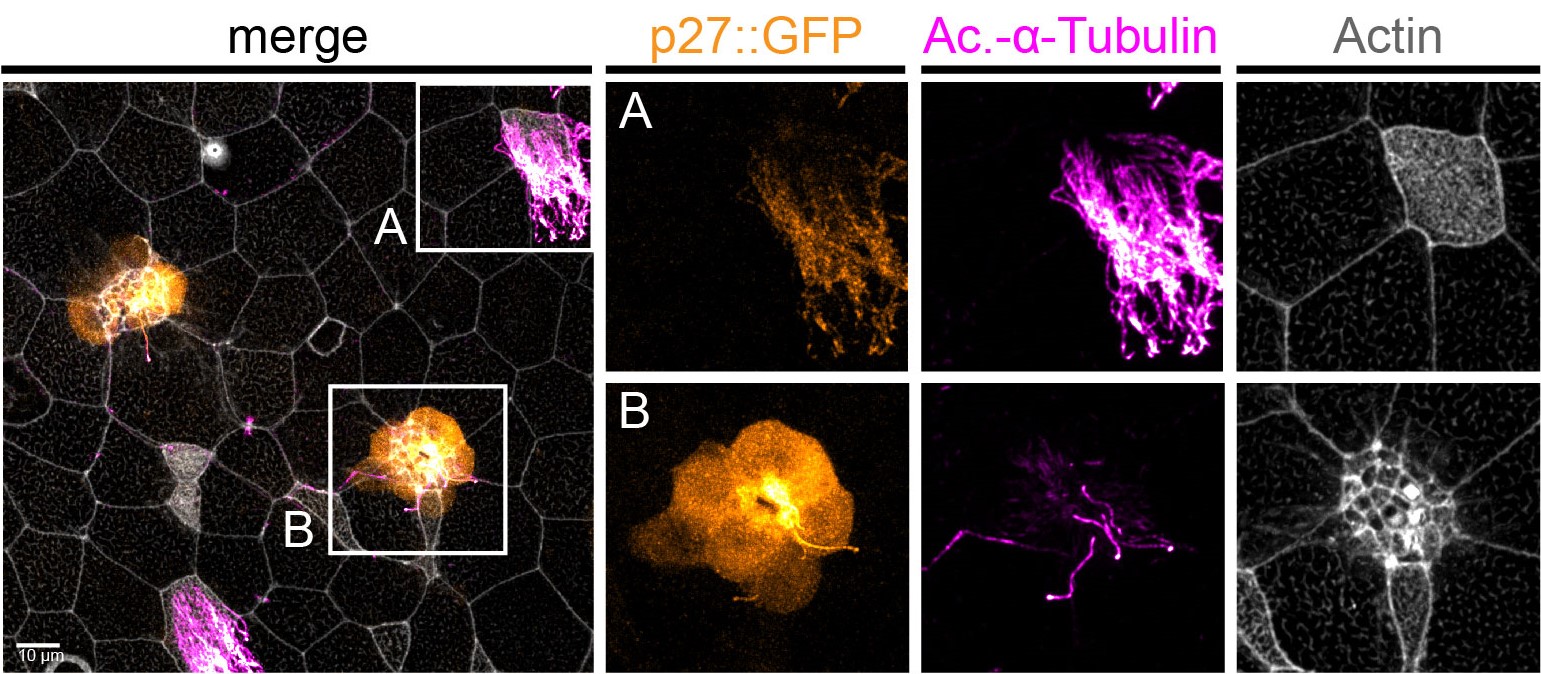
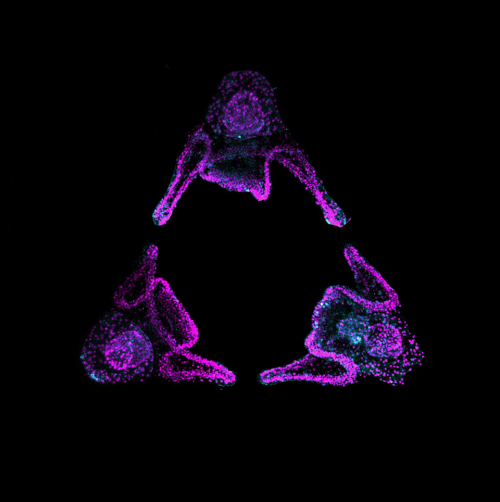
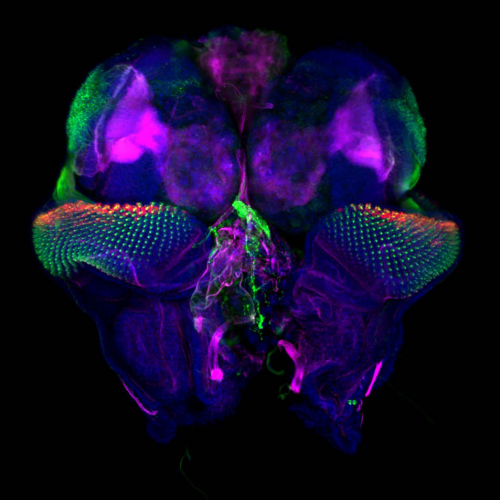
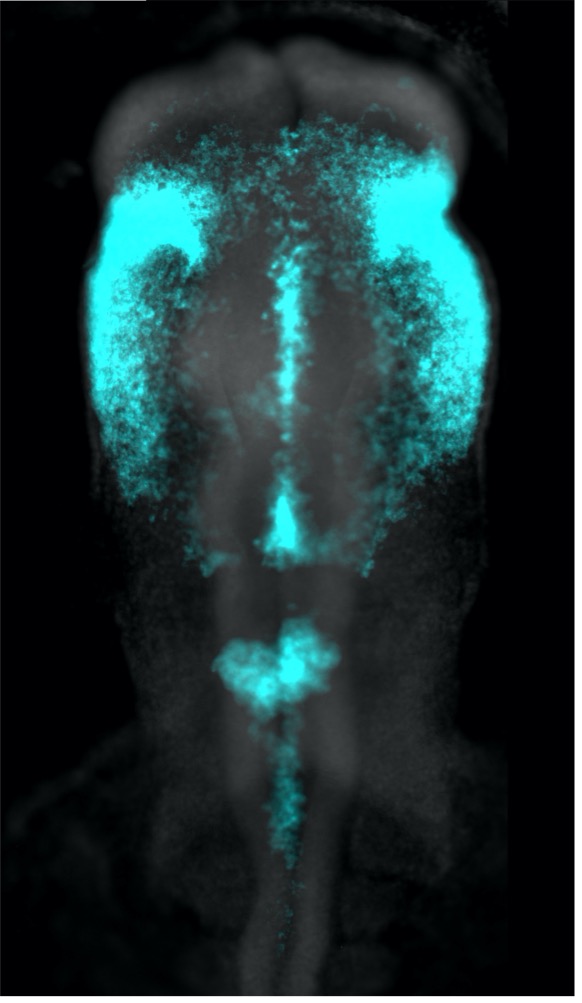
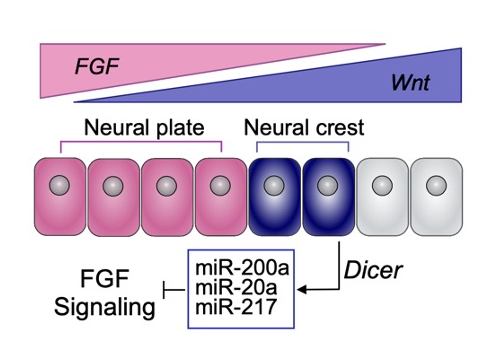
Figure 1 : MCCs are lost locally around the neuromasts of the lateral line. A)MCC stained for acetylated-α-tubulin and actin. MCC express low level of p27::GFP. B) Neuromast of the lateral line labeled by p27::GFP and stained for acetylated-α-tubulin and actin.
To understand the temporalrelationship between the migration of the lateral line primordium, NM deposition and the loss of MCCs, I started to transplant fluorescently labeled lateral line primordium cells into non-fluorescent hosts, and to use a transgenic reporter line (p27::GFP) (Rubbini et al., 2015) which expresses GFP in the lateral line primordium and NMs. Interestingly, I found that MCCs are still present while the primordium is migrating, but are lost when NMs emerge through intercalation in the epithelium (Figure 1). In parallel, I conducted immunofluorescent staining, confocal microscopy and analyzed scanning EM images, which showed that MCCs could be shed from the epithelium, suggesting removal through apoptosis. Therefore, I stained tadpoles with an anti-cleaved Caspase 3 antibody and performed TUNEL assays that showed signals exclusively in MCCs. This confirmed that MCCs over the lateral line were lost via apoptosis. Thus, we hypothesized that emergence of neuromasts induces loss of MCCs via shedding-apoptosis.
As we like to do in the lab, I first performed an easy and fast experiment to provide a proof-of-concept for our hypothesis that NM emergence is really the cause of local MCC loss. For that, I simply ablated the anterior part of the embryo where the lateral line primordium originates from and from where primordial cells migrate out in various directions to populate head, trunk and tail with NMs. This experiment confirmed that MCCs were not locally lost in absence of NM deposition.
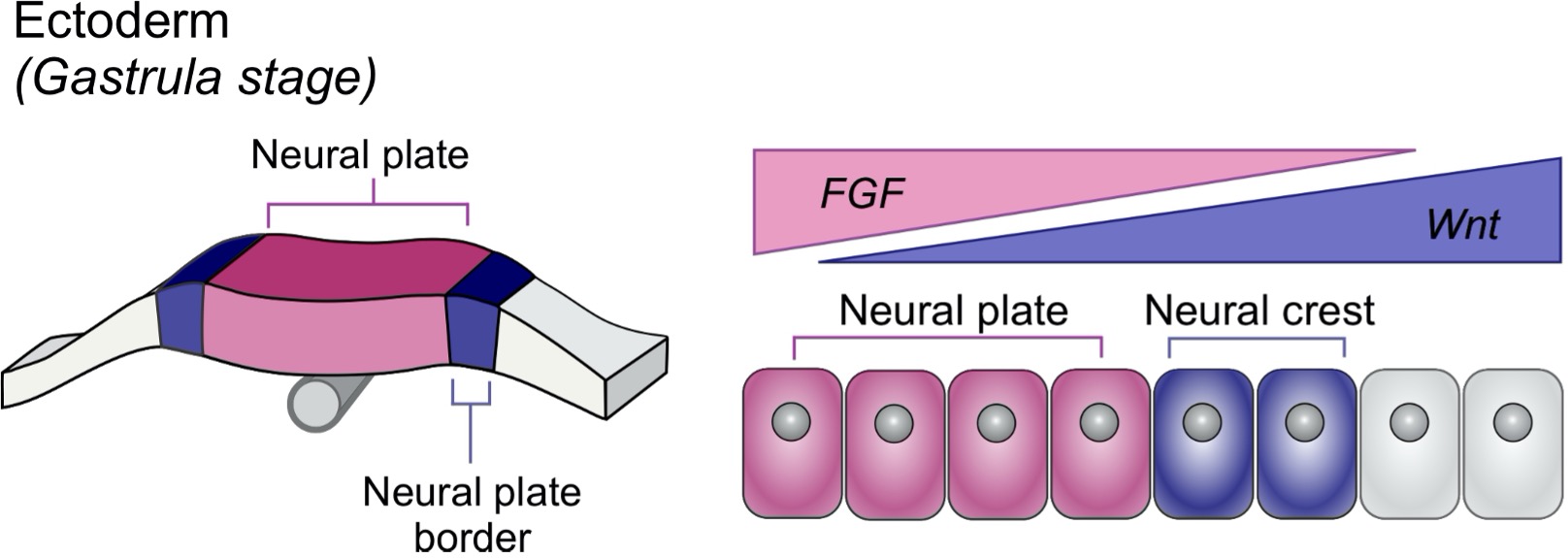
But how did NMs induce this loss of MCCs? Looking into the literature, we realized that NMs are signaling centers that express Notch ligands. During specification, MCCs are inhibited by Notch signaling, and mature MCCs retain some level of Notch receptor expression, which means that they could also respond to Notch signaling changes. This led us to hypothesize that high Notch signaling from NMs could signal to MCCs and induce apoptosis. It did not take too long to find out which ligands are expressed in NMs, because fellow graduate student Magdalena Brislinger in the lab is working on Notch signaling and has analyzed the expression of all Notch ligands and receptors throughout early Xenopus development. On her beautiful images of sectioned tadpoles stained for Notch ligand expression, we found that the lateral line primordium and NMs express jag1 at high levels and induce hes1 expression in the overlying epithelial cells. This validated that NMs are Notch signaling centers that communicate with epidermal cells. By incubating the embryos in DAPT, which inhibits Notch signaling, and by performing Caspase 3 and TUNEL assays, I could show that MCC apoptosis and loss over the lateral line were suppressed in absence of Notch activation, confirming that Notch signaling is required for MCC loss via apoptosis.
The majority of MCCs coordinately trans-differentiates into Goblet secretory cells
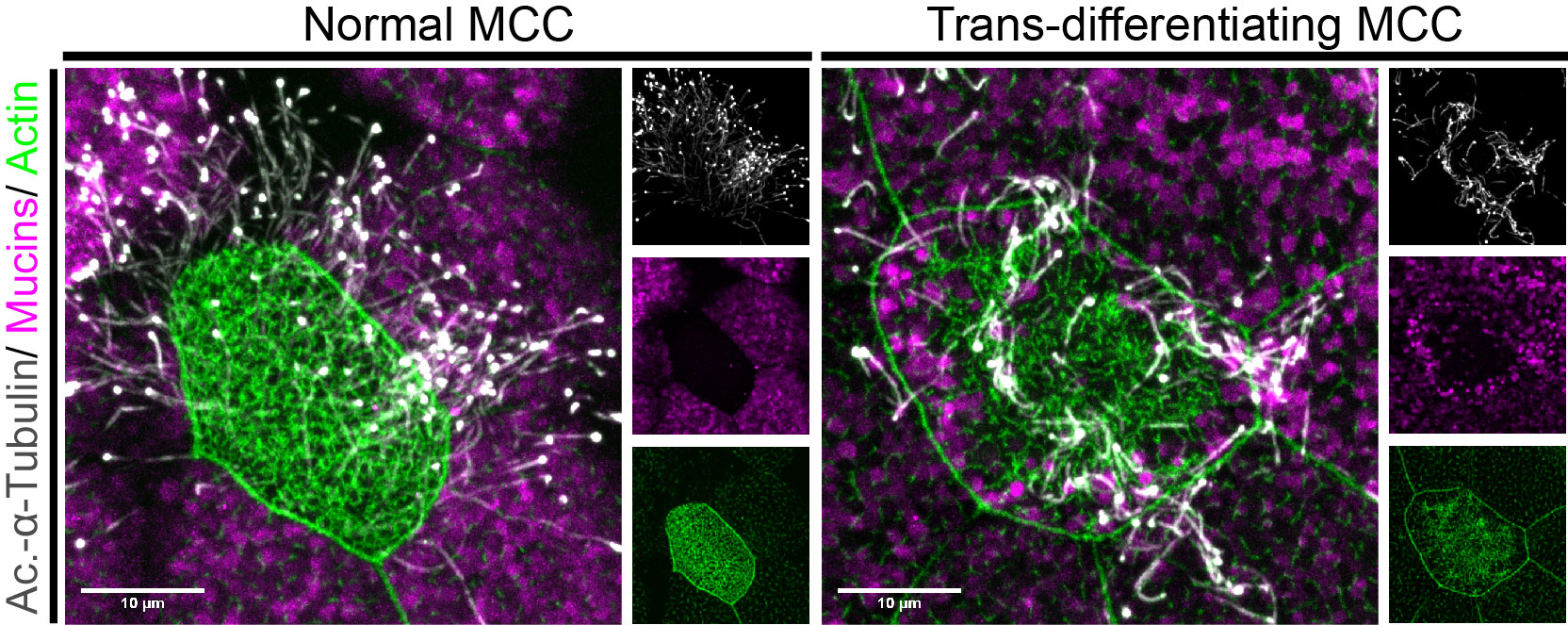
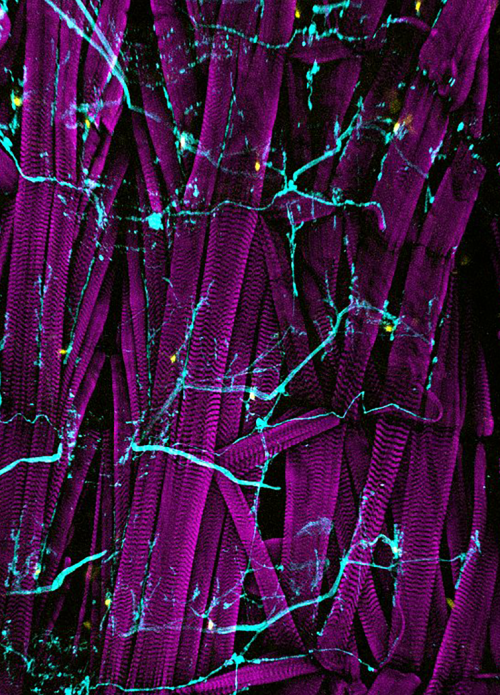
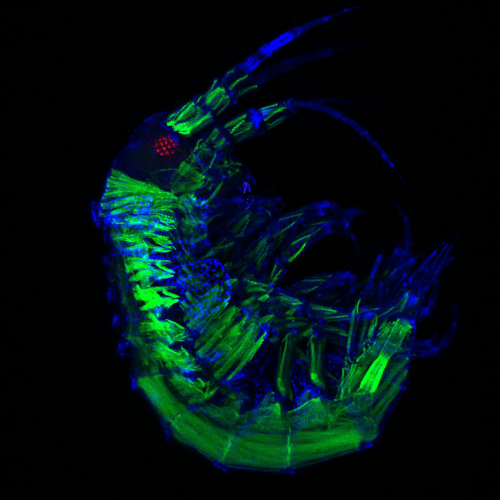
Figure 2: MCCs trans-differentiate into a mucous-secretory goblet cells. Left: Normal MCC stained for acetylated-α-tubulin (grey), PNA (magenta) and actin (green). Right: Trans-differentiating MCC stained for acetylated-α-tubulin (grey), PNA (magenta) and actin (green) shows reduce ciliation and acetylation as well as mucin production and apical actin remodeling.
But my investigations were not finished yet! Broad epidermal TUNEL staining was missing from areas farther away from the lateral line, which made us think that an alternative mode of MCC removal was used there. To find out what was going on, I stained tadpoles throughout the time of global MCC loss to visualize MCC cilia, to identify secretory cell types via mucus staining, and for F-actin to outline cell borders and to assess cell morphology. Interestingly, confocal microscopy on these samples revealed altered apical F-actin morphology in a subset of MCCs, which also stained positive for mucus. I will always remember the moment when I found those cells and, still new to the Xenopus field, I ask Peter naively if it was normal to see some MCCs with mucus, and he got all excited about the finding (Figure 2).Quantification of this dataset showed that while the overall number of MCCs decreased over time, the proportions of mucus-positive MCCs increased. This suggested MCC to goblet cell trans-differentiation as an additional mechanism for MCC removal in the Xenopus epidermis. Subsequently, we also found that mesoderm-derived intermediate Notch signaling levels cause MCC to goblet cell trans-differentiation, but only when thyroid hormone was produced, which elevated Jak/STAT signaling that has an anti-apoptotic effect and is required to allow MCCs to undergo this transition- probably by making them more resistant against stress. Based on these findings, a key aspect of the paper became the dual role of Notch in MCC apoptosis and cell fate change. We (and the reviewers of our paper) thought that a genetic manipulation of Notch signaling, which could induce both behaviors in young MCCs, would strongly support our statement. Thus, I wanted to use a Notch gain-of-function approach to manipulate MCCs specifically. So, I generated a construct that expresses constitutive active Notch intracellular domain NICD fused to GFP under the control of a MCC-specific promoter. The cloning seemed easy but not if you consider the unexpected magic of cloning. After struggling for weeks to have this construct ready and perform my last experiments for this paper, I finally succeeded to generate the construct and open a bottle of Champaign to celebrate my success. After injecting the construct, I could see nuclear GFP in MCCs, but importantly, a significant proportion of GFP-positive cells showed goblet cell morphology, demonstrating that Notch signaling activation in MCCs can trigger fate change. Additionally, TUNEL assays showed the induction of apoptosis in early stage tadpoles. Together, these experiments provide evidence that ectopic Notch signaling can induce apoptosis as well as cell fate conversion in MCCs.
MCCs retract cilia and loose basal body components
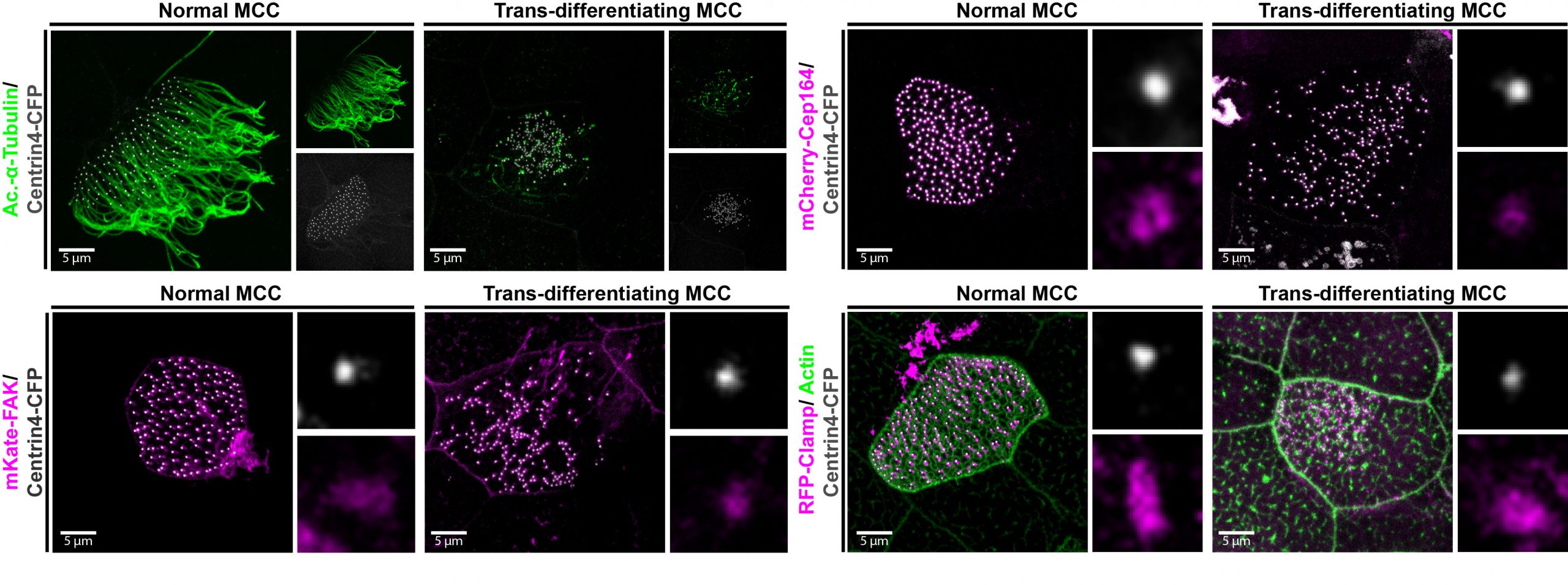
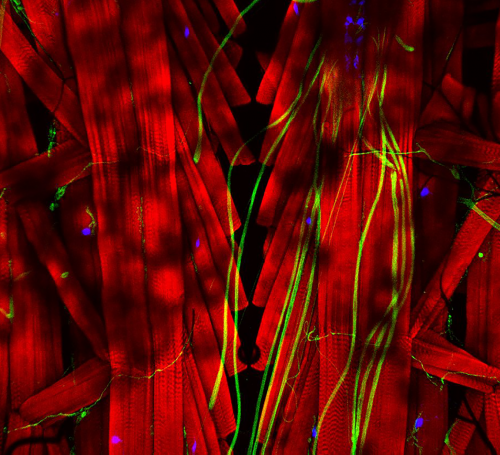
Figure 3: Trans-differentiating MCCs remodel basal body distribution and composition. Confocal micrograph of a normal MCC and a trans-differentating MCC reveals disorganized basal bodies (Centrin4-CFP, grey), cilia de-acetylation (Ac.-α-tubulin, green), F-actin remodeling (Actin, green) and reduce levels of basal body distal appendages proteins (mCherry-Cep164, magenta), actin interactors (FAK-RFP, magenta), and rootlet components (Clamp-RFP, magenta).
We also found that trans-differentiation is initiated through loss of ciliary gene expression, including foxj1 (a master transcription factor for motile cilia maintenance) and pcm1 (a protein that protects cilia and basal bodies from degradation). At the cellular level, we could observe altered proteostasis, cilia retraction, basal body elimination (Figure 3, Figure 4G) as well as initiation of mucus production and secretion. Some of these changes resembled processes observed during primary cilia retraction, which is initiated in cycling cells when they re-enter the cell cycle to divide. So, I wondered if MCCs that trans-differentiate and become goblet cells could they also re-enter the cell cycle and divide again? I found that trans-differentiating MCCs also lost expression of the cell cycle inhibitor p27, supporting the idea that MCCs could re-enter the cell cycle, and presence of a hybrid cilium (Liu et al, 2020) could suggest that MCCs retrain a parental centriole that could serve as a base for mitotic division (Figure 4E). Therefore, I tried to follow individual cells using live-cell imaging and various techniques to label individual MCCs before trans-differentiation, including photo-convertible proteins, MCC-specific fluorescent labeling, etc. However, either the constructs turned out to be toxic to the cells, or the labeling could not be restricted to individual MCCs, thus, I could not exclude the possibility that I followed co-converted goblet cells, or I lost the cells during imaging. So, setting up this experiment properly would require a transgenic line, which takes a long time to establish in the Xenopus laevis system. Sadly, due to these technical limitations, we were not able to provide genetic tracing data in this paper. But we are looking forward to find out more about the cellular behaviors of MCCs during and after the trans-differentiation process in the future. This will fill an extremely important gap in our understanding of tissue remodeling and MCC loss!
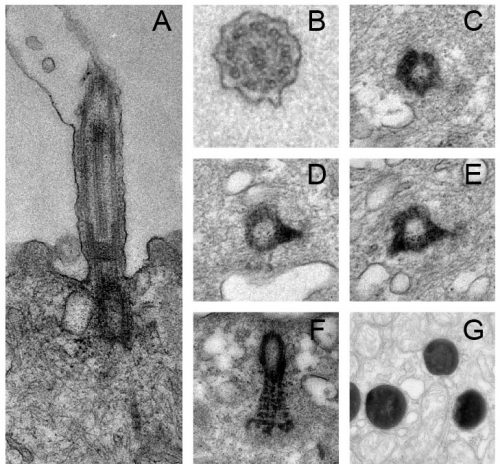
Figure 4: Cilia and basal body structure visualised by electron microscopy. A) Transversal section and transmission electron microscopy of a motile cilium and its associated basal body. Motile cilia of MCCs are composed microtubules in a 9+2 configuration, a transition zone and a basal body. B-F) Parallel sections and transmission electron microscopy of a cilium (B), basal bodies with one (D) or two (E) basal feet and rootlet (F). G) Trans-differentiating MCCs are enriched in electron-dense structures corresponding to lysosomes.
So, in summary, our work describes two modes for MCC loss during vertebrate development, the signaling regulation of these processes, and demonstrates that even cells with extreme differentiation features can undergo direct fate conversion (Tasca et al.,2021). In addition to our scientific findings, this project was an amazing experience for me personally. It is a fantastic feeling to know that I could unravel, in large parts, a decades-long mystery, and to generate insights into the molecular processes of mucociliary tissue remodeling. I also enjoyed the scientific investigation, the collaboration with group members as well as with the Mitchell lab, and with Martin Helmstädter from the group of Gerd Walz in our department, who provided the beautiful electron microscopy images for the paper.
References
Liu, Z., et al., Super-Resolution Microscopy and FIB-SEM Imaging Reveal Parental Centriole-Derived, Hybrid Cilium in Mammalian Multiciliated Cells. Dev Cell, 2020. 55(2): p. 224-236 e6. DOI: 10.1016/j.devcel.2020.09.016
Nishikawa, S., J. Hirata, and F. Sasaki, Fate of ciliated epidermal cells during early development of Xenopus laevis using whole-mount immunostaining with an antibody against chondroitin 6-sulfate proteoglycan and anti-tubulin: transdifferentiation or metaplasia of amphibian epidermis. Histochemistry, 1992. 98(6): p. 355-8. DOI: 10.1007/BF00271070
Rubbini, D., et al., Retinoic Acid Signaling Mediates Hair Cell Regeneration by Repressing p27kip and sox2 in Supporting Cells. J Neurosci, 2015. 35(47): p. 15752-66. DOI: 10.1523/JNEUROSCI.1099-15.2015
Smith, S.C., M.J. Lannoo, and J.B. Armstrong, Lateral-line neuromast development in Ambystoma mexicanum and a comparison with Rana pipiens. J Morphol, 1988. 198(3): p. 367-379. DOI: 10.1002/jmor.1051980310
Walentek, P. and I.K. Quigley, What we can learn from a tadpole about ciliopathies and airway diseases: Using systems biology in Xenopus to study cilia and mucociliary epithelia. Genesis, 2017. 55(1-2). DOI: 10.1002/dvg.23001
Tasca, A., et al., Notch signaling induces either apoptosis or cell fate change in multiciliated cells during mucociliary tissue remodeling. Dev Cell, 2021. 56(4): p. 525-539 e6. DOI: 10.1016/j.devcel.2020.12.005


 (2 votes)
(2 votes)