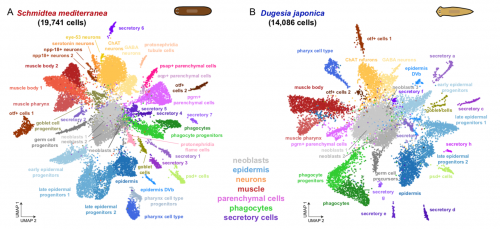
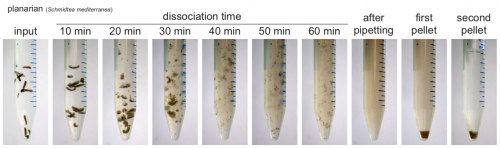
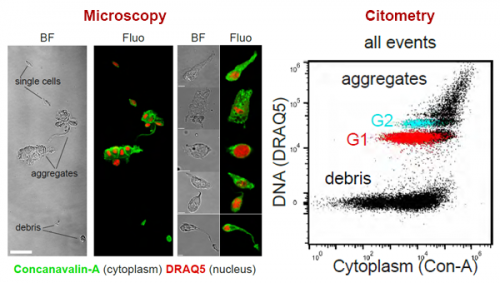
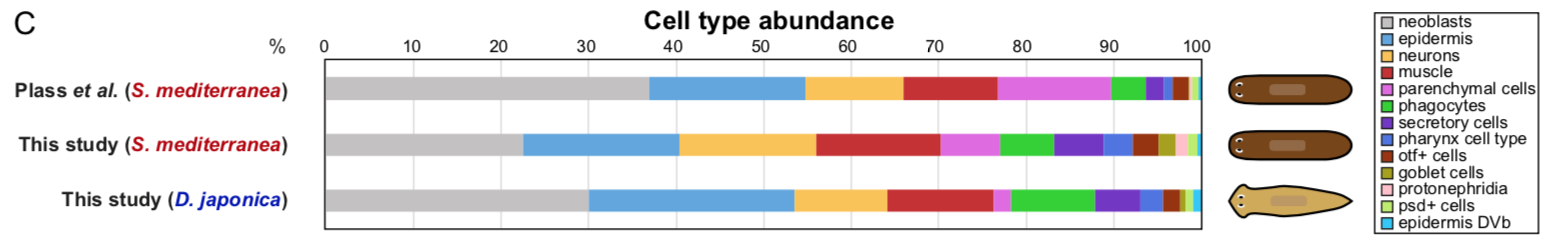
Back to bench is resetting my mental clock
Posted by Mayank Chugh, on 13 July 2020
My university has reopened and being reunited with the lab bench after months have reminded me again how much I love what I do for a living. Of course I enjoy the entire process of research from bench to publication, troubleshooting experiments and brainstorming ideas to presenting research stories. However, hands-on working is something I relish the most. It has a tangible and stimulatory effect on my experimental thought process and in general how I perceive and approach scientific questions.
Staying home for more than three months has been the largest break in my life since kindergarten and has affected my mental wellbeing. I had moved to Boston as a postdoc to study how forces shape tissue patterning and morphogenesis in zebrafish, after finishing PhD in single-molecule plant development in Tübingen, Germany. However, within two months of my arrival in Boston, Harvard stopped all the non-essential research due to soaring coronavirus pandemic.
Living pretty much alone in house arrest in a new city away from my partner and family has been extremely stressful and emotionally difficult. The new lab has been very supportive and connected throughout but the isolation – paired with the rejection from the postdoctoral fellowship I applied for – has given me an imposter syndrome. I felt the sadness growing, my moods shifting, confidence losing, and fear building in me. For example, during scientific zoom meetings with the lab and the department, I was not able to communicate my thoughts and ideas properly because being out of the lab and staying in isolation was toying with my state of mind. Of course, afterwards I then dealt with the guilt of it. The murder of George Floyd further added frustration and anger about the world we live in and how we treat each other. I could feel and acknowledge my worsening mental health. Therefore, I constantly talked about my feelings with my partner and my best friend who have been incredibly supportive irrespective of our separation by the continents. Although communication helps, I felt I still need tangible positivity to pull me out of the mental black hole. So, I have been taking time to reflect on positive things. For example, I have been excitedly rethinking about the scientific problems I am interested in and ways to approach them differently. And besides science, reading literary and historical fiction and baking cakes and pastries. I am sure I am not alone in experiencing such mental instability in this time. Each one of us is experiencing stress and anxiety in our own ways. What is most important is to acknowledge the symptoms and seek the help and support we need.
When I heard the university was reopening and we could go back to the lab, I was thrilled. This is exactly the positivity I think I have been longing for. The flipside to this positivity is that I was nervous about going back. I joined a zebrafish developmental and systems biology lab and had learned how to do crossings and microinjections in zebrafish right before the shutdown. Obviously, I have been anxious that I had forgotten the newly learned methods and whether I would be able to find reagents and consumables in the lab.
Now that I am in the lab again, of course, it is all the same as I knew it, physically. Except that the labs, corridors, hallways, bathrooms, meeting points, and common areas are labelled with safety distance reminders and are awfully silent and empty. There are no social hours or physical meetings. It feels like working late over Christmas. Even fish in the lab probably miss the hustle-bustle of colleagues. I, on the other hand, am delighted to be back in the lab and to see a few familiar masked faces around. Foremost, it is getting me back into the workday routine and breaking the monotony of alone and ‘relaxed’ working from home. More importantly, I am thrilled to be working on the bench again. I think there is an eternal joy that we wet-lab scientists find in bench work. To get back into the familiar working spirit, I resumed by molecular cloning experiments, which I am most comfortable with. Seeing bacterial colonies on the agar plate the day after bacterial transformation have already started to diminish my feelings of being an imposter. Turns out, I can still cross zebrafish and pull needles for microinjections. I am starting to find purpose again, after all work is the reason I moved to the US.

Although we are working in spatially and temporally coordinated lab shifts, the motivation to be in the lab and conduct experiments is more than enough for me to be feeling mentally healthy again. I understand it is not the ideal situation for any of us. But neither is the uncertainty of normality. I think by embracing little steps towards normality, we can overcome the mental hurdles posed by the pandemic. I realise my mental clock has started to shift towards normal: being back at the bench is definitely the sweeter side of our bitter-sweet labcoming experience.


 (11 votes)
(11 votes) (No Ratings Yet)
(No Ratings Yet)
 (1 votes)
(1 votes)

 FocalPlane
FocalPlane In this episode, supported by the Medical Research Council, we discover how researchers are letting the light shine in, literally, by bringing discoveries about the underlying genetic faults that cause eye diseases all the way through to game-changing clinical trials of gene therapy designed to save sight.
In this episode, supported by the Medical Research Council, we discover how researchers are letting the light shine in, literally, by bringing discoveries about the underlying genetic faults that cause eye diseases all the way through to game-changing clinical trials of gene therapy designed to save sight.